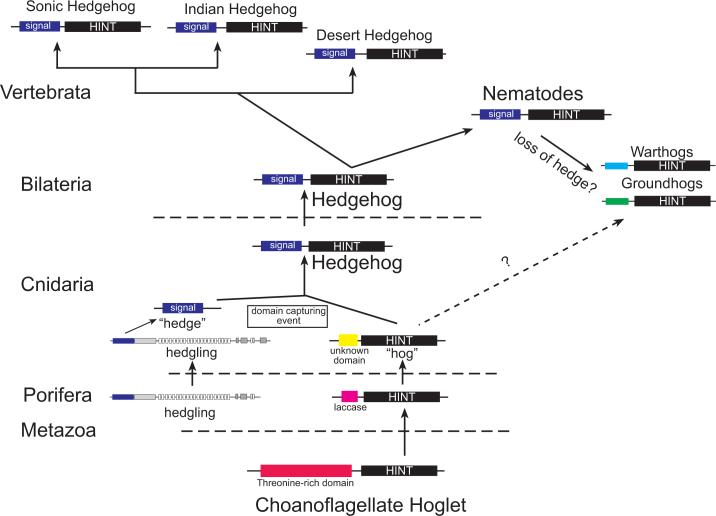
Figure 7. A model for Hedgehog evolution.
Data from the genomes of the anthozoan cnidarian Nematostella vectensis and the demosponge, Amphimedon queenslandica suggest that the origin of the Hedgehog signaling molecule arose in the eumetazoan ancestor (Adamska et al. 2007). The choanoflagellate, Monosiga ovata possesses a hint/intein “hog” containing gene (Snell et al., 2006), but no gene possessing a signal or “hedge” domain has been found. The demosponge, A. queenslandica possesses both hint/intein genes as well as a gene possessing a “hedge” domain tethered to a fat-like cadherin (Adamska et al. 2007). An ortholog of this gene has been found in the N. vectensis genome. However, N. vectensis also possesses two true hedgehogs as well as three hint/intein genes. While Hint/intein containing genes have not been identified in most bilaterians surveyed, C. elegans, which lacks true hedgehog genes (Aspock et al., 1999; Hao et al., 2006a), possesses two families of Hint/intein containing genes, with novel N-terminal domains (warthog and groundhog). However, a true hedgehog gene has been identified in the slowly evolving nematode, Xiphinema index (M. Blaxter pers. comm.). Based on phylogenetic analyses (Fig. 2) it appears likely that nematode groundhog, warthog, and quahog genes are derived from an ancestral nematode true hedgehog gene (solid arrow), however additional data may be necessary to rule out the possibility that these genes could also be derived from non-bilaterian hint-only genes (dashed arrow) The hedgehog ligands diversified within the Vertebrata.