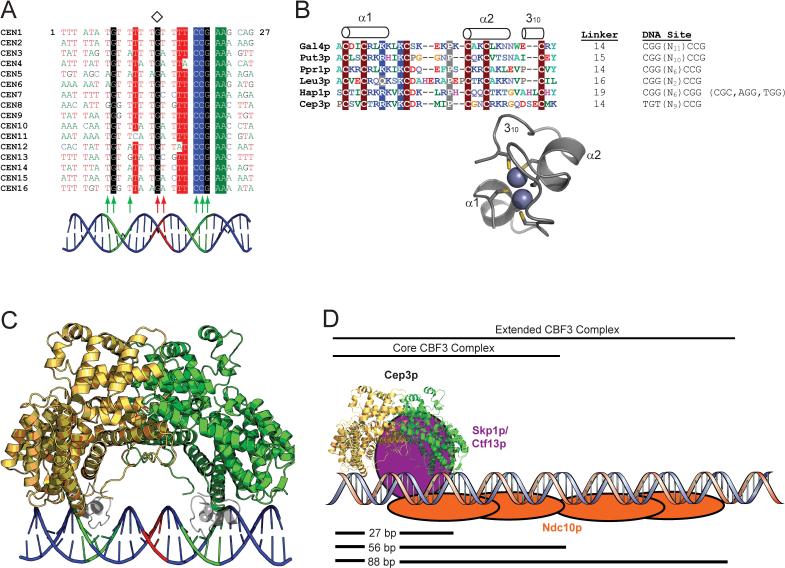
Figure 4. A model of CBF3-DNA assembly.
A. Multiple sequence alignment of the first 27 bases of the 56-bp DNAse I footprint of CDEIII from the 16 S. cerevisiae centromeres. The center of pseudosymmetry at the conserved base 14G is denoted by a diamond. Highly conserved positions are shaded. The positions of Cep3p (green) and Ctf13p (red) protein-DNA crosslinks (Espelin et al., 1997) are indicated by arrows. A B-DNA model of CDEIII is shown in cartoon form, with the putative Cep3p half-sites in green and the putative Ctf13p major groove binding site in red. B. The Zn2Cys6 cluster domain. Multiple sequence alignment of Zn2Cys6 cluster domains from six S. cerevisiae proteins, with conserved residues shaded and secondary structural elements indicated above the alignment. The length of the linker connecting the Zn2Cys6 cluster with αA (in Cep3p) or with the coiled-coil dimerization element (in the other proteins), is indicated, along with the DNA sites recognized by each protein. The Zn2Cys6 cluster from Hap1p (PDB ID 1WHT) is presented as a ribbon diagram with the Zn atoms and Cys sidechains shown (King et al., 1999). The two-residue insertion between Cys5 and Cys6 that forms the 310 helix in Hap1p is also present in Cep3p. C. A model for Cep3p-CDEIII binding. Zn2Cys6 clusters from the Hap1p structure have been docked into the two Cep3p half-sites on CDEIII. The twofold axis of Cep3p and the pseudo twofold axis of CDEIII have been superimposed. The polarity of the left-half site is ambiguous; this model approximates what the Cep3p-CDEIII complex could look like if the left half-site forms an inverted repeat (as in Gal4p) retaining overall two-fold symmetry. D. Model of CBF3-CDEIII Assembly. Ctf13p must bind asymmetrically to the Cep3p dimer and contact CDEIII in the major groove halfway between the Cep3p half-sites, as well as link Cep3p to Ndc10p. The Skp1p-Ctf13p heterodimer (purple) has been placed in an arbitrary position that fulfills those constraints. Ndc10p (orange) is modeled to reflect DNA crosslinking and electrophoretic mobility shift data, which suggest that one Ndc10p dimer binds the first 56 bases of CDEIII (as part of the core CBF3 complex), and a second Ndc10p dimer binds the region between 56 and 88 bp (to form the extended CBF3 complex) (Espelin et al., 1997).