Abstract
Background
Variability in hypothalamic-pituitary-adrenal (HPA) axis activity has been shown to be influenced by genetic factors and related to great metabolic differences such as obesity. The aim of this study was to investigate molecular bases of genetic variability of the adrenal sensitivity to ACTH, a major source of variability, in Meishan (MS) and Large White (LW) pigs, MS being reported to exhibit higher basal cortisol levels, response to ACTH and fatness than LW. A pig cDNA microarray was used to identify changes in gene expression in basal conditions and in response to ACTH stimulation.
Results
Genotype and/or ACTH affected the expression of 211 genes related to transcription, cell growth/maintenance, signal transduction, cell structure/adhesion/extra cellular matrix and protein kinase/phosphatase activity. No change in the expression of known key regulator proteins of the ACTH signaling pathway or of steroidogenic enzymes was found. However, Mdh2, Sdha, Suclg2, genes involved in the tricarboxylic acid (TCA) pathway, were over-expressed in MS pigs. Higher TCA cycle activity in MS than in LW may thus result in higher steroidogenic activity and thus explain the typically higher cortisol levels in MS compared to LW. Moreover, up-regulation of Star and Ldlr genes in MS and/or in response to ACTH suggest that differences in the adrenal function between MS and LW may also involve mechanisms requisite for cholesterol supply to steroidogenesis.
Conclusion
The present study provides new potential candidate genes to explain genetic variations in the adrenal sensitivity to ACTH and better understand relationship between HPA axis activity and obesity.
Background
The hypothalamic-pituitary-adrenal (HPA) axis and more particularly the adrenal gland constitute a principal node of the mammalian endocrine system. The main function of the adrenal cortex is to produce glucocorticoids (cortisol in pig) and mineralocorticoids under the influence of pituitary adrenocorticotropic hormone (ACTH). Adrenal hormones, essential for survival, play important roles in stress responses, metabolism regulation, immunity, reproduction, water and salt balance and various brain functions [1].
Large variations in HPA axis activity (i.e. basal and in response to stress) have been related to genetic factors (in human [2], in mice [3], in rats [4-6], in pigs [7] and in birds [8-10]). Variations in HPA axis activity are also related to important metabolic differences. For example, human abdominal obesity has been associated with alterations in HPA axis functioning [11,12]. In a recent experiment comparing five genetic stocks of pigs (Large White, Landrace, Duroc, Meishan and Piétrain), a positive relationship between cortisol levels in urine (basal and after transportation stress) and body fat content was found both within and across breeds [13]. In addition, Meishan pigs have been reported to exhibit higher basal cortisol levels, response to ACTH and body fat content than Large White pigs [7,14]. Meishan and Large White lines of pigs thus constitute a valuable biological model to investigate the relationship between HPA axis activity and metabolic regulation.
Among the different genetic mechanisms involved in HPA axis activity variability [15], several experimental findings suggest that sensitivity to ACTH is a major target in human [16] and in rats [4,17]. In the pig, genetic-based differences in cortisol secretion were shown in response to corticotropin-releasing hormone (CRH) although the ACTH response did not differ among individuals [18]. Moreover, metabolic clearance of cortisol bears no relationship to the cortisol response to ACTH [19]. Previous findings [14] indicate that the difference in HPA axis activity between LW and MS pigs may originate from the adrenal gland although these breeds also differ in corticosteroid-binding globulin (CBG) levels that influence circulating levels of cortisol [20]. In this study, we explore the molecular mechanism responsible for the difference in adrenal sensitivity to ACTH in MS and LW pigs.
The actions of ACTH in the adrenal cortex are mediated via two temporally distinct pathways. Acute and chronic regulation of steroidogenesis occur within minutes and hours, respectively [21,22]. The acute response is initiated by the mobilization and delivery of the substrate, cholesterol, for steroid hormone biosynthesis from the outer to the inner mitochondrial membrane, where it is metabolized to pregnenolone by the cytochrome P450 cholesterol side chain cleavage enzyme (P450scc) [23]. The slower, long-lasting response to ACTH directs transcription of genes encoding the steroidogenic enzymes [22,24]. Most studies of ACTH action have focused on the long term induction (several hours) of transcripts either by investigating transcripts with specific functions in steroidogenesis [22,24] or by genome-wide analysis [25-27]. However, the acute response to ACTH stimulation has been reported to require de novo protein synthesis (for review see ref. [21]). The steroidogenic acute regulatory protein (StAR), that is responsible for the transfer of cholesterol from the outer to the inner mitochondrial membrane, has been proposed to be the rate-limiting and regulated step in steroidogenesis [23,28]. Changes in Star gene expression induced by ACTH have been observed as early as 30 min and were maximally elevated between 1 and 3 h [29]. On the other hand, cortisol responses to ACTH injections in Meishan and Large White pigs have been reported to be at maximum levels at 1 hour [14,30]. Taking together these findings, we hypothesized that transcriptional regulation may in fact take place in the acute response to ACTH, i.e. within 1 hour, in both lines of pigs. Differences in transcriptional regulation at the adrenal gland could already exist between both genotypes in basal conditions because basal cortisol levels are typically greater in MS pigs.
Considering the findings described above and the literature concerning acute regulation of transcripts by ACTH in adrenals, we undertook a microarray analysis of gene expression in the adrenal glands of MS and LW pigs under basal conditions and in response to acute stimulation by ACTH. The aim of this study was to investigate the molecular bases of the genetic differences in adrenal sensitivity to ACTH. We found that genotype and/or ACTH affected the expression of 211 genes which provide new potential candidate genes to explain genetic variations in the adrenal sensitivity to ACTH.
Results
Cortisol levels
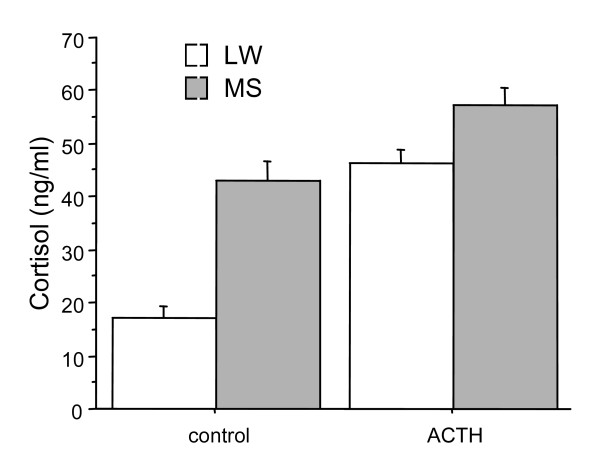
Comparison of plasma cortisol concentrations (figure 1) in control and ACTH-injected pigs showed significant breed (p < 0.0001) and treatment (p < 0.0001) effects and significant interaction (p ≤ 0.001). In control animals, basal cortisol levels were higher in Meishan pigs than in Large White pigs. Injection of ACTH increased cortisol levels in both genotypes but to a larger extent in Large White than in Meishan pigs.
Figure 1.
Cortisol concentrations (ng/ml plasma). Cortisol concentrations (ng/ml plasma) measured in Large White (LW) and Meishan (MS) piglets either untreated (control) or 1 hour after injection (ACTH) of a high dose (250 μg per animal) of 1–24 ACTH (Immediate synacthen). (n = 6 per experimental group, means ± SE).
Gene transcript regulation by genotype and ACTH treatment
Using the normalization process described in the methods section, 3496 of the 8959 transcripts present on the pig cDNA array (approximately 40%) were found to be expressed in adrenal glands in our experimental conditions. Using the criterion of < 5% FDR, 241 transcripts were identified to be significantly up-regulated or downregulated by genotype and/or ACTH challenge (see additional file 1). Among these, 211 transcripts corresponded to unique annotated transcripts (five present twice) and 25 remained unknown. These 241 transcripts were categorized according to the genotype and/or treatment effect and ordered by absolute fold change. Table 1 lists transcripts most significantly affected by genotype and/or treatment at p < 0.0001. Among these, 51 transcripts were significantly affected by genotype (39 transcripts up-regulated and 12 down regulated in Meishan), 21 transcripts were significantly affected by ACTH treatment (12 transcripts up-regulated and six down regulated in response to ACTH treatment) and 36 transcripts were significantly affected by both genotype and ACTH treatment. The major functional categories for these genes included transcription regulation, cell growth/maintenance, signal transduction, structural/cell adhesion/ECM and protein kinase activity.
Table 1.
Genes differentially expressed depending on genotype and/or ACTH treatment
| Gene symbol | Gene name | Gene Function | p-value (ANOVA) | Fold Change | |||||
| genotype: MS/LW | treatment: ACTH/control | ||||||||
| genotype | treatment | interaction | control | ACTH | LW | MS | |||
| Genes up-regulated in Meishan pigs | |||||||||
| Rnf2 | ring finger protein 2 | Transcription regulation | 2.81E-13 | 7.89E-01 | 1.96E-02 | 3.5 | 2.5 | 1.2 | -1.2 |
| Mdk | midkine (neurite growth-promoting factor 2) | Cell growth/maintenance | 4.97E-06 | NS | NS | 2.0 | 1.5 | 1.2 | -1.1 |
| Snf1lk | SNF1-like kinase | Protein Kinase activity | 8.19E-05 | NS | NS | 1.8 | 1.8 | 1.2 | 1.3 |
| Ccnd1 | cyclin D1 | Cell growth/maintenance | 9.83E-06 | NS | NS | 1.7 | 1.5 | 1.1 | 1.0 |
| Cd99 | CD99 molecule | Structural/Cell adhesion/ECM | 1.01E-05 | NS | NS | 1.7 | 1.6 | 1.0 | 1.0 |
| Col21a1 | collagen, type XXI, alpha 1 | Structural/Cell adhesion/ECM | 9.39E-05 | NS | NS | 1.7 | 1.5 | -1.0 | -1.2 |
| Gprc5b | G protein-coupled receptor, family C, group 5, member B | Signal transduction (glutamatergic) | 9.53E-07 | NS | NS | 1.6 | 1.5 | 1.1 | -1.0 |
| Ldhd | lactate dehydrogenase D | ATP biosynthesis | 3.77E-06 | NS | NS | 1.6 | 1.6 | 1.1 | 1.1 |
| Ppp1r1a | protein phosphatase 1, regulatory (inhibitor) subunit 1A | Phosphatase inhibitor | 8.18E-06 | NS | NS | 1.6 | 1.4 | -1.0 | -1.2 |
| Mrps6 | mitochondrial ribosomal protein S6 | Translation | 1.25E-06 | NS | NS | 1.6 | 1.4 | 1.1 | -1.1 |
| Traf5 | TNF receptor-associated factor 5 | Signal transduction | 5.80E-05 | NS | NS | 1.5 | 1.5 | 1.0 | 1.0 |
| Cse1l | CSE1 chromosome segregation 1-like (yeast) | Protein transport | 2.98E-06 | NS | NS | 1.5 | 1.4 | 1.1 | 1.0 |
| Hist2h2aa | histone cluster 2, H2aa4 | Nucleosome Assembly | 2.10E-05 | NS | NS | 1.5 | 1.5 | -1.1 | -1.1 |
| Cd99 | CD99 molecule | Structural/Cell adhesion/ECM | 2.27E-05 | NS | NS | 1.5 | 1.6 | -1.1 | 1.0 |
| Mif | macrophage migration inhibitory factor (glycosylation-inhibiting factor) | Cell growth/maintenance | 1.25E-05 | NS | NS | 1.5 | 1.3 | 1.1 | -1.0 |
| Ptpmt1 | protein tyrosine phosphatase, mitochondrial 1 | Protein Phosphatase activity | 1.73E-08 | NS | NS | 1.5 | 1.5 | 1.0 | 1.1 |
| Mdh2 | malate dehydrogenase 2, NAD (mitochondrial) | Tricarboxylic acid cycle | 2.59E-06 | NS | NS | 1.5 | 1.5 | 1.0 | 1.1 |
| Malat1 | metastasis associated lung adenocarcinoma transcript 1 (non coding RNA) | unknown | 4.07E-05 | NS | NS | 1.5 | 1.3 | -1.0 | -1.2 |
| Suclg2 | succinate-CoA ligase, GDP-forming, beta subunit | Tricarboxylic acid cycle | 1.82E-05 | NS | NS | 1.4 | 1.3 | 1.0 | -1.1 |
| Eif3s7 | eukaryotic translation initiation factor 3, subunit 7 zeta, 66/67kDa | Translation regulation | 4.61E-05 | 5.87E-01 | 2.64E-02 | 1.4 | 1.1 | 1.1 | -1.2 |
| Magi3 | membrane associated guanylate kinase, WW and PDZ domain containing 3 | Protein Kinase activity | 2.40E-05 | NS | NS | 1.4 | 1.8 | -1.2 | 1.0 |
| Ppap2b | phosphatidic acid phosphatase type 2B | Protein Phosphatase activity | 3.45E-05 | 6.74E-02 | 4.48E-02 | 1.4 | 2.2 | -1.0 | 1.5 |
| Cys1 | cystin 1 | unknown | 1.08E-08 | NS | NS | 1.4 | 1.5 | -1.1 | -1.0 |
| Txnl2 | thioredoxin-like 2 | Electron transport | 5.63E-05 | NS | NS | 1.4 | 1.3 | 1.0 | 1.0 |
| C8orf53 | chromosome 8 open reading frame 53 | unknown | 1.34E-05 | NS | NS | 1.4 | 1.4 | 1.1 | 1.1 |
| C14orf2 | chromosome 14 open reading frame 2 | unknown | 1.65E-05 | NS | NS | 1.4 | 1.3 | 1.0 | -1.0 |
| Dusp26 | dual specificity phosphatase 26 (putative) | unknown | 3.78E-06 | NS | NS | 1.4 | 1.4 | 1.0 | 1.0 |
| Sod2 | superoxide dismutase 2, mitochondrial | Super oxide metabolism | 1.61E-06 | NS | NS | 1.4 | 1.5 | -1.1 | -1.0 |
| Ssr3 | signal sequence receptor, gamma (translocon-associated protein gamma) | Cotranslational membrane targeting | 7.82E-05 | NS | NS | 1.4 | 1.3 | 1.1 | 1.0 |
| Xpnpep1 | X-prolyl aminopeptidase (aminopeptidase P) 1, soluble | Proteolysis | 3.05E-05 | NS | NS | 1.3 | 1.3 | -1.0 | -1.0 |
| Hint1 | histidine triad nucleotide binding protein 1 | Signal transduction | 6.24E-05 | NS | NS | 1.3 | 1.2 | 1.1 | 1.0 |
| Hsbp1 | heat shock factor binding protein 1 | Transcription regulation | 6.13E-05 | NS | NS | 1.3 | 1.2 | 1.0 | -1.0 |
| Mrps10 | mitochondrial ribosomal protein S10 | Structural constituent of ribosome | 6.05E-05 | NS | NS | 1.3 | 1.3 | -1.0 | 1.0 |
| Lpp | LIM domain containing preferred translocation partner in lipoma | unknown | 4.71E-05 | NS | NS | 1.2 | 1.3 | 1.1 | 1.1 |
| Pgk1 | phosphoglycerate kinase 1 | Protein Kinase activity | 1.83E-05 | NS | NS | 1.2 | 1.4 | -1.1 | 1.1 |
| Ppap2d | phosphatidic acid phosphatase type 2d | Protein Phosphatase activity | 6.15E-06 | NS | NS | 1.2 | 1.3 | -1.0 | 1.1 |
| Atp5b | ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide | ATP biosynthesis | 1.07E-05 | NS | NS | 1.2 | 1.1 | 1.0 | -1.0 |
| Ccny | cyclin Y | unknown | 1.59E-05 | NS | NS | 1.2 | 1.3 | 1.0 | 1.1 |
| Cox6b1 | cytochrome c oxidase subunit Vib polypeptide 1 (ubiquitous) | Oxydoreductase activity | 9.02E-05 | NS | NS | 1.2 | 1.3 | -1.0 | 1.1 |
| Genes down-regulated in Meishan pigs | |||||||||
| C1qa | complement component 1, q subcomponent, A chain | Cell-cell signaling | 3.08E-06 | NS | NS | -2.1 | -1.5 | -1.1 | 1.2 |
| Cited1 | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 | Transcription regulation | 1.88E-10 | NS | NS | -1.8 | -2.0 | 1.1 | -1.0 |
| Tmem14c | transmembrane protein 14C | Structural/Cell adhesion/ECM (mito) | 4.24E-06 | 4.81E-01 | 4.54E-02 | -1.8 | -1.3 | -1.2 | 1.1 |
| Hmgb3 | high-mobility group box 3 | Transcription regulation | 2.00E-08 | NS | NS | -1.6 | -1.4 | -1.2 | -1.0 |
| Tmem14c | transmembrane protein 14C | Structural/Cell adhesion/ECM (mito) | 5.40E-09 | 3.37E-01 | 1.61E-02 | -1.6 | -1.3 | -1.1 | 1.1 |
| Bphl | biphenyl hydrolase-like (serine hydrolase; breast epithelial mucin-associated antigen) | Amino acid metabolism | 2.71E-06 | NS | NS | -1.5 | -1.4 | -1.1 | -1.0 |
| Ttll12 | tubulin tyrosine ligase-like family, member 12 | unknown | 8.52E-08 | NS | NS | -1.5 | -1.6 | 1.1 | -1.0 |
| Gpx3 | glutathione peroxidase 3 (plasma) | Peroxidase activity | 8.06E-08 | NS | NS | -1.5 | -1.5 | -1.0 | -1.1 |
| Fkbp4 | FK506 binding protein 4, 59kDa | Protein binding | 4.64E-05 | NS | NS | -1.4 | -1.3 | 1.0 | 1.1 |
| Gns | glucosamine (N-acetyl)-6-sulfatase (Sanfilippo disease IIID) | Glycosaminoglycan catabolism | 5.48E-05 | NS | NS | -1.3 | -1.5 | 1.1 | -1.1 |
| Nr1h2 | nuclear receptor subfamily 1, group H, member 2 | Transcription regulation | 1.30E-05 | NS | NS | -1.2 | -1.3 | 1.0 | -1.1 |
| H2afj | H2A histone family, member J | Nucleosome Assembly | 3.50E-05 | NS | NS | -1.2 | -1.3 | -1.0 | -1.1 |
| Genes up-regulated by ACTH | |||||||||
| Bag3 | BCL2-associated athanogene 3 | Cell growth/maintenance | 3.76E-01 | 9.34E-11 | 5.19E-03 | 1.2 | -1.3 | 3.0 | 2.0 |
| Ier3 | immediate early response 3 | Cell growth/maintenance | 4.49E-01 | 2.83E-09 | 3.81E-02 | 1.3 | -1.1 | 2.6 | 1.9 |
| Adamts1 | ADAM metallopeptidase with thrombospondin type 1 motif, 1 | Cell growth/maintenance | NS | 2.37E-06 | NS | 1.4 | -1.1 | 2.6 | 1.7 |
| Gadd45a | growth arrest and DNA-damage-inducible, alpha | Regulation of protein kinase activity | NS | 1.46E-07 | NS | 1.1 | -1.1 | 2.1 | 1.7 |
| Id3 | inhibitor of DNA binding 3, dominant negative helix-loop-helix protein | Transcription regulation | NS | 4.69E-07 | NS | -1.1 | -1.2 | 2.0 | 1.9 |
| Tfpi2 | tissue factor pathway inhibitor 2 | Structural/Cell adhesion/ECM | NS | 4.72E-07 | NS | 1.1 | 1.0 | 1.8 | 1.7 |
| Odc1 | ornithine decarboxylase 1 | Cell growth/maintenance | NS | 2.85E-06 | NS | 1.2 | 1.0 | 1.6 | 1.4 |
| Tomm20 | translocase of outer mitochondrial membrane 20 homolog (yeast) | Protein transport (mb ext mito) | NS | 9.51E-05 | NS | 1.2 | 1.1 | 1.5 | 1.4 |
| Timp1 | TIMP metallopeptidase inhibitor 1 | Cell growth/maintenance | NS | 2.88E-07 | NS | 1.0 | -1.1 | 1.4 | 1.3 |
| Ddx3x | DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked | RNA helicase activity | NS | 1.19E-05 | NS | 1.1 | 1.0 | 1.4 | 1.3 |
| C10orf46 | chromosome 10 open reading frame 46 | unknown | NS | 6.70E-06 | NS | 1.0 | 1.2 | 1.4 | 1.6 |
| Gnl2 | guanine nucleotide binding protein-like 2 (nucleolar) | unknown | NS | 3.73E-05 | NS | 1.1 | -1.0 | 1.3 | 1.2 |
| Genes down regulated by ACTH | |||||||||
| Stat5a | signal transducer and activator of transcription 5A | Transcription regulation | NS | 8.12E-05 | NS | -1.0 | 1.0 | -1.4 | -1.3 |
| Wasf2 | WAS protein family, member 2 | Signal transduction (G-protein) | NS | 7.81E-05 | NS | -1.0 | 1.0 | -1.3 | -1.3 |
| Unc84b | unc-84 homolog B (C. elegans) | Cell growth/maintenance | NS | 4.26E-05 | NS | 1.2 | -1.0 | -1.3 | -1.6 |
| Cntf | ciliary neurotrophic factor | Signal transduction | NS | 4.93E-05 | NS | -1.0 | 1.1 | -1.3 | -1.2 |
| Nfe2l2 | nuclear factor (erythroid-derived 2)-like 2 | Transcription regulation | NS | 5.59E-06 | NS | 1.1 | 1.0 | -1.3 | -1.3 |
| Pcbd1 | pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) | Transcription regulation | NS | 5.70E-05 | NS | 1.1 | 1.0 | -1.2 | -1.3 |
| Bmpr2 | bone morphogenetic protein receptor, type II (serine/threonine kinase) | Protein Kinase activity | NS | 3.15E-05 | NS | -1.1 | 1.0 | -1.2 | -1.1 |
| Ctnnb1 | catenin (cadherin-associated protein), beta 1, 88kDa | Transcription regulation | NS | 1.01E-05 | NS | -1.0 | -1.1 | -1.2 | -1.2 |
| Gdf8 | growth differentiation factor 8 | Signal transduction (TGFb) | NS | 5.41E-05 | NS | 1.1 | -1.1 | -1.2 | -1.3 |
| Genes upregulated in Meishan and by ACTH | |||||||||
| Crem | cAMP responsive element modulator | Transcription regulation | 4.34E-02 | 2.70E-10 | NS | 1.7 | 1.1 | 4.4 | 3.0 |
| Cd83 | CD83 molecule | Signal transduction | 6.77E-06 | 3.80E-10 | NS | 1.9 | 1.5 | 3.2 | 2.6 |
| Eif1b | eukaryotic translation initiation factor 1B | Translation regulation | 9.08E-06 | 7.89E-10 | NS | 1.5 | 1.9 | 2.5 | 3.3 |
| Tob1 | transducer of ERBB2, 1 | Transcription regulation | 4.11E-02 | 6.96E-08 | NS | 1.1 | 1.0 | 2.5 | 2.3 |
| Ldlr | low density lipoprotein receptor (familial hypercholesterolemia) | Cholesterol mobilization | 5.19E-03 | 4.15E-07 | NS | 1.6 | 1.3 | 2.3 | 1.8 |
| Chchd2 | coiled-coil-helix-coiled-coil-helix domain containing 2 | unknown | 4.72E-04 | 3.36E-06 | NS | 1.4 | 1.3 | 1.6 | 1.5 |
| Ckb | creatine kinase, brain | Structural/Cell adhesion/ECM | 1.03E-06 | 3.00E-02 | NS | 2.1 | 1.9 | 1.3 | 1.2 |
| Mki67ip | MKI67 (FHA domain) interacting nucleolar phosphoprotein | rRNA metabolism | 1.25E-04 | 9.14E-05 | NS | 1.3 | 1.3 | 1.3 | 1.3 |
| Maob | monoamine oxidase B | Electron transport | 4.43E-05 | 4.23E-03 | NS | 1.4 | 1.3 | 1.2 | 1.2 |
| Dnaja | DnaJ (Hsp40) homolog, subfamily A, member 2 | Cell growth/maintenance | 1.59E-05 | 2.55E-03 | NS | 1.5 | 1.2 | 1.2 | 1.0 |
| H19 | H19, imprinted maternally expressed untranslated mRNA | unknown | 9.02E-07 | 5.79E-03 | NS | 1.5 | 1.6 | 1.1 | 1.3 |
| Dleu1 | deleted in lymphocytic leukemia, 1 | Cell growth/maintenance | 4.90E-07 | 2.14E-03 | NS | 1.5 | 2.0 | 1.1 | 1.4 |
| Genes up-regulated in Meishan and down-regulated by ACTH | |||||||||
| Selenbp1 | selenium binding protein 1 | Transporter activity | 2.98E-05 | 1.41E-02 | NS | 1.3 | 1.5 | -1.3 | -1.1 |
| C16orf33 | chromosome 16 open reading frame 33 | unknown | 5.03E-05 | 3.98E-02 | NS | 1.2 | 1.3 | -1.2 | -1.0 |
| Ech1 | enoyl Coenzyme A hydratase 1, peroxisomal | Fatty acid metabolism | 5.27E-07 | 6.31E-03 | NS | 1.4 | 1.4 | -1.2 | -1.2 |
| Anpep | alanyl (membrane) aminopeptidase (aminopeptidase N, aminopeptidase M, microsomal aminopeptidase, CD13, p150) | Development/Morphogenesis | 5.65E-12 | 8.27E-03 | NS | 2.1 | 1.9 | -1.1 | -1.2 |
| Iscu | iron-sulfur cluster scaffold homolog (E. coli) | unknown | 6.09E-05 | 1.03E-03 | NS | 1.3 | 1.2 | -1.1 | -1.2 |
| Enpp2 | ectonucleotide pyrophosphatase/phosphodiesterase 2 (autotaxin) | Signal transduction (G-protein) | 8.22E-05 | 6.17E-03 | NS | 1.4 | 1.3 | -1.1 | -1.2 |
| Klhl22 | kelch-like 22 (Drosophila) | unknown | 2.75E-04 | 9.94E-05 | NS | 1.3 | 1.1 | -1.1 | -1.2 |
| Sdha | succinate dehydrogenase complex, subunit A, flavoprotein (Fp) | Tricarboxylic acid cycle | 3.38E-05 | 3.08E-02 | NS | 1.2 | 1.2 | -1.0 | -1.1 |
| Genes down regulated in Meishan and by ACTH | |||||||||
| Ctdsp2 | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 | unknown | 1.84E-02 | 4.68E-08 | NS | -1.1 | -1.3 | -1.6 | -1.8 |
| Pecam1 | platelet/endothelial cell adhesion molecule (CD31 antigen) | Signal transduction | 2.74E-05 | 8.45E-04 | NS | -1.6 | -1.6 | -1.4 | -1.5 |
| Dab2 | disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila) | Cell growth/maintenance | 2.92E-02 | 5.63E-05 | NS | -1.1 | -1.3 | -1.4 | -1.7 |
| Cyb5b | cytochrome b5 type B (outer mitochondrial membrane) | Transporter activity | 4.15E-03 | 2.59E-06 | NS | -1.1 | -1.3 | -1.4 | -1.6 |
| Ostf1 | osteoclast stimulating factor 1 | Signal transduction | 1.66E-07 | 1.08E-02 | NS | -1.7 | -1.5 | -1.3 | -1.1 |
| Gdf8 | growth differentiation factor 8 | Signal transduction (TGFb) | 4.95E-03 | 6.71E-05 | NS | -1.1 | -1.3 | -1.3 | -1.4 |
| Nfat5 | nuclear factor of activated T-cells 5, tonicity-responsive | Transcription regulation | 2.75E-03 | 1.27E-05 | NS | -1.2 | -1.2 | -1.3 | -1.3 |
| Acox1 | acyl-Coenzyme A oxidase 1, palmitoyl | Fatty acid metabolism | 7.62E-10 | 4.31E-04 | NS | -1.7 | -1.5 | -1.2 | -1.1 |
| Ephx1 | epoxide hydrolase 1, microsomal (xenobiotic) | Proteolysis | 1.60E-06 | 2.62E-02 | NS | -1.8 | -1.6 | -1.2 | -1.1 |
| Ccdc85b | coiled-coil domain containing 85B | unknown | 1.66E-05 | 4.54E-02 | NS | -1.6 | -1.4 | -1.2 | -1.1 |
| A2m | alpha-2-macroglobulin | Protein transport | 2.45E-10 | 3.68E-03 | NS | -1.8 | -1.7 | -1.2 | -1.1 |
| Nono | non-POU domain containing, octamer-binding | mRNA processing | 1.01E-06 | 3.80E-03 | NS | -1.4 | -1.3 | -1.2 | -1.1 |
| Ifltd1 | intermediate filament tail domain containing 1 | unknown | 6.75E-06 | 1.81E-04 | NS | -1.2 | -1.4 | -1.2 | -1.3 |
| Myst4 | MYST histone acetyltransferase (monocytic leukemia) 4 | Transcription regulation | 9.48E-06 | 1.18E-02 | NS | -1.3 | -1.3 | -1.1 | -1.2 |
| Genes down regulated in Meishan and up-regulated by ACTH | |||||||||
| Gadd45b | growth arrest and DNA-damage-inducible, beta | Regulation of protein kinase activity | 6.38E-04 | 1.41E-11 | NS | -1.3 | -1.3 | 2.4 | 2.4 |
| C14orf4 | chromosome 14 open reading frame 4 | unknown | 8.66E-03 | 7.41E-05 | NS | -1.0 | -1.2 | 1.4 | 1.2 |
Genes differentially expressed between Meishan (MS) and Large White (LW) pigs and/or in response to acute stimulation by ACTH were identified using a pig cDNA microarray made up of 8959 cDNA clones. The most significant affected genes (p < 0.0001) were listed in the table, categorized on the basis of up or down-regulated by genotype and/or ACTH and sorting by decreasing fold changes. (unknown transcripts were excluded) Complete list is given in the additional file 1.
Quantitative analysis
Eleven genes highly significantly affected by genotype and/or treatment factors were selected for further examination by quantitative real-time PCR (Table 2). Changes in transcripts levels were confirmed for nine genes. Correlation coefficients between expression levels as measured with membrane hybridization and real-time PCR were significant (r2 between 0.51 and 0.90, p < 0.05) for 9 out of 11 genes tested (see additional file 2). The magnitudes of the changes were roughly similar between the array and the real-time PCR except for the largest fold-changes (higher than 2 fold) that seemed dampened on the microarrays. An exception was Rnf2 gene, which showed no difference between genotypes of pigs or in response to ACTH when assayed by quantitative RT-PCR, but which exhibited a significant genotype difference when tested against the pig cDNA array. This disparity may have resulted from a false-discovery error or from cross-hybridization of transcripts to region of similarity in the arrayed Rnf2 cDNA. The second exception was Fxc1, which showed no significant difference when assayed by quantitative RT-PCR, but which exhibited a significant genotype difference when tested against the pig cDNA array. However, the fold changes between breeds found on the array were of low magnitude (< 1.5-fold).
Table 2.
Measurement of genotype and/or ACTH effects on transcript accumulation by relative quantitative real-time PCR and comparison with microarray data
| Gene symbol | Microarray | Real time PCR | ||||||||||||
| p-value (ANOVA) | Fold Change | p-value (ANOVA) | Fold Change | |||||||||||
| genotype: MS/LW | treatment: ACTH/control | genotype: MS/LW | treatment: ACTH/control | |||||||||||
| genotype | treatment | interaction | control | ACTH | LW | MS | genotype | treatment | interaction | control | ACTH | LW | MS | |
| Rnf2 | 2.81E-13 | 7.89E-01 | 1.96E-02 | 3.5 | 2.5 | 1.2 | -1.2 | 0.82 | 0.64 | 0.81 | 1.1 | -1.0 | 1.1 | 1.0 |
| Anpep | 5.65E-12 | 8.27E-03 | NS | 2.1 | 1.9 | -1.1 | -1.2 | < 0.0001 | 0.040 | 0.056 | 3.9 | 3.0 | -1.0 | -1.4 |
| Gadd45b | 6.38E-04 | 1.41E-11 | NS | -1.3 | -1.3 | 2.4 | 2.4 | 0.028 | < 0.0001 | 0.33 | -1.5 | -1.5 | 2.6 | 2.6 |
| Eif1b | 9.08E-06 | 7.89E-10 | NS | 1.5 | 1.9 | 2.5 | 3.3 | 0.009 | < 0.0001 | 0.040 | 1.4 | 1.9 | 2.9 | 4.1 |
| Crem | 4.34E-02 | 2.70E-10 | NS | 1.7 | 1.1 | 4.4 | 3.0 | 0.18 | < 0.0001 | 0.43 | 1.6 | 1.3 | 6.4 | 5.4 |
| A2m | 2.45E-10 | 3.68E-03 | NS | -1.8 | -1.7 | -1.2 | -1.1 | < 0.0001 | 0.11 | 0.54 | -2.1 | -2.1 | -1.3 | -1.2 |
| Ldlr* | 5.19E-03 | 4.15E-07 | NS | 1.6 | 1.3 | 2.3 | 1.8 | ND | ND | ND | ND | ND | ND | ND |
| Ckb | 1.03E-06 | 3.00E-02 | NS | 2.1 | 1.9 | 1.3 | 1.2 | 0.004 | 0.43 | 0.88 | 2.3 | 2.1 | 1.3 | 1.2 |
| Dab2 | 2.92E-02 | 5.63E-05 | NS | -1.1 | -1.3 | -1.4 | -1.7 | 0.34 | 0.052 | 0.63 | -1.3 | -1.2 | -1.8 | -1.6 |
| Ldlr* | 1.50E-02 | 1.14E-03 | 1.06E-02 | 1.5 | 1.0 | 1.6 | 1.1 | 0.069 | < 0.0001 | 0.80 | 1.4 | 1.2 | 2.2 | 2.0 |
| Fxc1 | 3.06E-04 | 2.35E-02 | NS | 1.2 | 1.3 | 1.1 | 1.1 | 0.28 | 0.16 | 0.48 | -1.1 | -1.4 | 1.6 | 1.2 |
| Star | 4.85E-04 | NS | NS | 1.6 | 1.2 | 1.4 | 1.1 | 0.053 | 0.26 | 0.406 | 1.4 | 1.1 | 1.3 | 1.0 |
| Mc2r | ND | ND | ND | ND | ND | ND | ND | 0.95 | 0.24 | 0.77 | -1.1 | 1.0 | 1.2 | 1.4 |
| Scarb1 | NS | NS | NS | NS | NS | NS | NS | 0.89 | 0.25 | 0.75 | 1.1 | -1.0 | 1.2 | 1.1 |
| Bzrp | ND | ND | ND | ND | ND | ND | ND | 0.017 | 0.64 | 0.089 | -2.3 | -1.2 | -1.4 | 1.4 |
| Sqle | NS | NS | NS | NS | NS | NS | NS | 0.16 | 0.39 | 0.32 | -1.1 | -1.5 | 1.3 | -1.0 |
| Cyp11a1 | NS | NS | NS | NS | NS | NS | NS | 0.85 | 0.36 | 0.37 | 1.2 | -1.1 | 1.4 | 1.0 |
| Hsd3b1 | NS | NS | NS | NS | NS | NS | NS | 0.51 | 0.17 | 0.94 | -1.1 | -1.1 | -1.3 | -1.3 |
| Cyp21 | NS | NS | NS | NS | NS | NS | NS | 0.022 | 0.90 | 0.33 | -1.2 | -1.5 | 1.1 | -1.1 |
| Cyp11b | NS | NS | NS | NS | NS | NS | NS | 0.13 | 0.27 | 0.94 | 1.3 | 1.3 | 1.2 | 1.2 |
| Per2 | ND | ND | ND | ND | ND | ND | ND | 0.46 | 0.040 | 0.066 | -1.2 | 1.8 | -2.3 | -1.0 |
| Cry1 | NS | NS | NS | NS | NS | NS | NS | 0.96 | 0.48 | 0.62 | 1.1 | -1.1 | 1.2 | 1.0 |
| Bmal1 | ND | ND | ND | ND | ND | ND | ND | 0.002 | 0.21 | 0.70 | -2.5 | -2.9 | -1.4 | -1.6 |
The changes in expression levels for 11 genes shown by microarray analysis to be significantly regulated by genotype and/or ACTH were quantified by real-time PCR. For comparison, relative expression values for each gene were determined in each animal (fold change, n = 6 pigs per experimental point). Changes in expression levels were also analyzed for 11 additional genes of great interest for our study whereas they were either shown by microarray analysis to be not significant (NS, p > 0.05) or not determined (ND) since they were not included on the microarray or due to the data normalization process. * LDLR gene was represented by 2 cDNA clones on the microarray and changes in its expression were verified by real-time PCR using only the cDNA sequence for which exons/introns structure could be deduced.
Eleven additional genes involved in ACTH signaling, metabolism and mobilization of cholesterol, steroidogenesis and clock genes that were either not found to be significantly affected by genotype or ACTH treatment (Scarb1, Sqle, Cyp11a1, Hsd3b1, Cyp21, Cyp11b, Cry1) or not present on the array (Mc2r, Bzrp, Per2, Bmal1) were selected for further examination by quantitative real-time PCR because they were interesting for our study (Table 2). The absence of significant changes in the microarray study was confirmed for six out of seven genes.
The Mc2r gene that encodes the ACTH receptor was not present on the array. Quantitative real-time PCR showed that the Mc2r gene was not significantly affected by genotype or by ACTH treatment. Inasmuch as the present data showed that two transcripts involved in cholesterol transport (i.e. Ldlr and Star) were significantly affected by genotype and ACTH treatment, changes in the expression of Scarb1 and Bzrp genes, also involved in cholesterol transport, were measured by quantitative RT-PCR. Quantitative RT-PCR analysis showed that genotype and ACTH treatment did not affect the expression of Scarb1. However, the expression of Bzrp gene was found to be greater in Large White pigs whereas Bzrp was not found expressed on the array probably due to the low expression level of this gene and to the data normalization process used in the microarray analysis. Sqle is a key gene involved in an early step of cholesterol biosynthesis de novo. Its expression was not significantly affected by genotype or ACTH on the array. This was confirmed by real-time PCR. Cyp11a1, Hsd3b1, Cyp21 and Cyp11b that encode enzymes involved in steroidogenesis were unaffected by genotype or ACTH on the microarrays. Quantitative RT-PCR confirmed that neither genotype nor ACTH affected the levels of those genes except for Cyp21. While no significant difference was found when tested against the pig cDNA array, expression of Cyp21 was found to be greater in LW pigs when assayed by quantitative RT-PCR, but the fold changes were of low magnitude. Finally, 3 clock genes Per2, Cry1 and Bmal1 reported to be involved in circadian corticosteroids biosynthesis or adrenal responsiveness to ACTH were further investigated by quantitative real-time PCR. No significant change in Cry1 expression on the microarray was confirmed by real-time PCR. The two 2 other clock genes, Per2 and Bmal1, not present on the microarray were found significantly affected by ACTH treatment and genotype, respectively. Per2 was down-regulated in response to ACTH treatment and expression of Bmal1 was greater in LW pigs.
Discussion
We used a comprehensive gene expression profiling by microarray analysis to identify groups of genes differentially expressed by genotype and/or by acute ACTH treatment. This is the first gene array analysis investigating in vivo adrenal response to an acute ACTH stimulation and exploring genetic variability at the adrenal level by using different breeds of pigs (i.e. an interesting biological model because pigs produce cortisol as humans). In microarrays, which included almost 8960 transcripts, the present results indicate that genotype and/or ACTH treatment affected the levels of 211 genes in adrenals. Moreover, although previous gene array analyses of ACTH action have been conducted in vitro and/or focused on the effects of chronic stimulation [25,27], our experiments demonstrate in vivo that acute ACTH treatment affects a large number of transcripts.
For the vast majority of affected transcripts, the changes were less than 2-fold excepted for some, probably of great interest, for which changes were as much as 4.5-fold. While some studies reported that small changes may be due to the tendency of microarray analysis to underestimate fold changes in transcripts accumulation [31], comparison of fold changes between microarray and real-time PCR analysis showed the accuracy of nylon microarrays used, even if fold-changes higher than 2-fold seemed to be somewhat underestimated. Our results suggest that genotypic difference and ACTH action may produce relatively small changes in transcript accumulation but these small changes could well be of physiological significance [32].
The dose of ACTH used in our study has been shown to maximally activate cortisol production within one hour [14,30] and thus, may produce maximum effects on steroidogenesis. Cortisol concentrations measured in our study were consistent with previous results reporting that cortisol levels induced by ACTH were higher in Meishans than in Large Whites, as well as basal cortisol levels [7,14]. However, ACTH did not affect gene expression of steroidogenesis enzymes (i.e. Cyp11a1, Cyp17, 3βhsd, Cyp21, Cyp11b). This result is consistent with the fact that acute stimulation by ACTH had little effect on adrenal P450s and steroidogenic enzymes while in contrast, long-term ACTH treatment provokes profound changes in the mRNA levels of many adrenal steroidogenic enzymes [22,29,33]. Interestingly, no differences in expression of steroidogenesis genes were found between Meishan and Large White pigs. These results suggest that the difference of corticosteroidogenesis between Meishan and Large White pigs is not triggered by changes in gene expression of adrenal P450 and 3βHSD enzymes under basal state or following acute ACTH stimulation. Nevertheless, we cannot exclude that steroidogenic activity might be higher in Meishan pigs than in Large White pigs. Indeed, expression of several genes (Mdh2, Sdha and Suclg2) involved in the tricarboxylic acid (TCA) pathway was greater in MS pigs. The main catalytic function of TCA cycle is to provide reducing equivalents to the respiratory complexes [34], for example, steroid hydroxylation [35]. Moreover, the TCA cycle also contributes to the synthesis of heme [34], necessary for the prosthetic groups of the steroidogenic cytochrome P450s [36]. In this respect, it is worth noting that Alas1, the rate-limiting enzyme in heme biosynthesis [37], shows a greater expression in MS than in LW pigs (see additional file 1). These mechanisms may together result in a higher steroidogenic activity by supplying more reducing equivalents and heme to steroidogenic enzymes. This hypothesis is supported by previous observations indicating that heme availability limited adrenal corticosteroid biosynthesis [38] and by recent data showing that acute stimulation of steroid production by ACTH was significantly increased when heme oxygenase activity was inhibited [39].
The effects of ACTH are mediated through the ACTH receptor (MC2R) belonging to the melanocortin receptor family (MCRs). The binding of ACTH to its cognate G protein-coupled receptor promotes the activation of protein kinase A and MAPK-dependent signaling cascades that collectively initiate adrenal-specific steroidogenic transcriptional programs [21,22,40]. Findings from numerous in vitro studies support the notion that ACTH is a positive regulator of ACTH-R mRNA expression [33,41,42]. More particularly, Winnay and Hammer [43] demonstrated in vitro that ACTH stimulation acutely activates the Mc2r gene promoter (i.e. within 80 min). In our study, we showed that ACTH did not affect Mc2r gene expression within 1 hour and that this gene was not differentially expressed between MS and LW pigs. Similarly, no difference in gene expression encoding key regulator proteins of the ACTH signaling pathway (i.e. G protein, Adenylate Cyclase, Protein Kinase A and MAPK ERK1, ERK2) was found in our study. On the other hand, the levels of Crem (cAMP response element modulator), a cAMP-dependent transcription factor that functions to activate genes involved in steroidogenesis [44], were increased in presence of ACTH and higher in MS than LW pigs. Moreover, trophic hormone stimulation of steroidogenic cells has been shown to result in the activation of G proteins that stimulate adenylate cyclase activity and produce increased intracellular levels of cAMP [21]. Thus, high levels of Crem in MS compared to LW and in response to ACTH may result to a larger increase of signal transduction induced by ACTH.
A constant supply of cholesterol is required within adrenal cells for steroidogenesis. The rate-limiting step for steroidogenesis is the movement of unesterified cholesterol into mitochondria where it can then be metabolized by CYP11A1 and other enzymes in the steroidogenic pathway (for review see ref. [45]). The movement of cholesterol into the mitochondria is mediated by steroidogenic acute regulatory protein (StAR) [23] and other partners such as peripheral-type benzodiazepine receptor or translocator protein [46]. Interestingly, greater expression of the Star gene in Meishan than in Large White pigs found in our study suggests that enhanced cholesterol transport into mitochondria may contribute to the higher corticosteroid biosynthesis found in MS pigs compared to LW pigs. All studies on StAR function agree that this enzyme mediates acute stimulation of steroidogenesis in response to ACTH administration and requires de novo protein synthesis (for review see ref. [21]). However, we did not find changes in transcript levels of Star 1 h following ACTH treatment while changes in Star gene expression induced by ACTH have been previously observed as early as 30 min and levels were maximally elevated between 1 and 3 h in the rat [29]. Nevertheless, phosphorylation of more preexisting StAR protein in MS pigs, a mechanism involved in the acute response to ACTH stimulation [47], could contribute to the higher cortisol levels induced in response to ACTH in MS than in LW pigs.
The unesterified cholesterol needed for steroidogenesis can be derived from several different sources (for review see ref. [45]). In our study we did not find differences in gene expression of enzymes involved in endogenous cellular cholesterol synthesis, such as Sqle or Hmgcr, between genotypes or in response to ACTH while those genes have been reported to be regulated by ACTH [25,27]. On the other hand, cellular cholesterol delivery for steroidogenesis includes uptake of lipoprotein-derived cholesterol via low density lipoprotein (LDL) receptor mediated endocytic pathways and SRB1 (Scavenger Receptor class B, type1) mediated "selective" pathways (for review see ref. [45]). Interestingly we found higher Ldlr expression in MS than in LW pigs and in response to ACTH but no changes were observed in Scarb1 expression.
Changes in Ldlr expression in response to ACTH found in our study are consistent with Ldlr up-regulation reported in vitro following 24 h ACTH treatment in Y1 mouse adrenal cells [25]. Conversely, while Scarb1 have also been reported to be up-regulated by 24 h ACTH treatment in Y1 mouse adrenal cells [25], and to a larger extent than Ldlr, we did not find change in Scarb1 expression. Our results indicate that receptor-mediated endocytic uptake of LDL-cholesterol may be a more important source of cholesterol for adrenal steroidogenesis in pigs as is the case in humans [48], while it appears to play a negligible role in mouse [45]. Moreover, this is the first evidence indicating in vivo that the acute response to ACTH may involve cellular cholesterol supply for steroidogenesis via Ldlr regulation.
A large number of genes found to be differentially expressed in our study encode transcription factors. Most of them have not been yet described as requisite in transcription networks involved in adrenal steroidogenesis. Nevertheless, they are potential interesting candidates, particularly those that were affected by both genotype and ACTH treatment. We were particularly interested in peripheral clock genes (such as Bmal1, Per2, Cry1), because recent studies reported that in the adrenal they regulate a large number of genes involved in general cellular processes (e.g. protein synthesis) as well as in pathways related to major organ-specific function (e.g. corticosteroid biosynthesis) and probably adjust adrenal sensitivity to ACTH [49-51]. Interestingly, while Per2 and Cry1 were not differentially expressed in our study, Bmal1 gene showed less expression in MS pigs. Thus, we can not exclude that differences in the expression level of some clock genes may be involved in differences in basal cortisol levels and adrenal reactivity to ACTH between MS and LW pigs. Further studies are needed to investigate other clock genes and to clarify how clock-controlled transcriptional rhythms in adrenals could contribute to the differences observed between both lines of pigs.
Phosphorylation and dephosphorylation mechanisms might be also involved in the difference of adrenal function between LW and MS pigs because a large number of differentially expressed genes encode diverse protein kinases and protein phosphatases. Among them Snf1lk (SIK1 protein) constitute a valuable candidate since it was reported to be an important regulator in the early phase of ACTH-signaling in the adrenal cortex [52].
Conclusion
In conclusion, we report differential gene expression in adrenal in two lines of pigs in basal conditions and following acute ACTH treatment. Some of the genes have been already reported to be implicated in adrenal physiology, but the majority has not been documented as directly involved in steroidogenesis regulation or as acutely ACTH-responsive. Although their contributions to adrenal function merit further investigation, we may speculate on the involvement of a few of them. The higher cortisol levels in basal state and in response to ACTH in MS than in LW pigs was probably not triggered by changes in gene expression of known key regulator proteins of the ACTH signaling pathway and steroidogenic enzymes. However, a higher TCA cycle activity in MS pigs than in LW pigs may explain the higher steroidogenic activity by supplying more reducing equivalents and heme to steroidogenic enzymes. Alternately, differences in the adrenal function between MS and LW pigs involve likely mechanisms requisite for cholesterol supply to steroidogenesis. The genes described in this report are thus excellent potential candidates to mediate the genetic differences in adrenal steroidogenesis, particularly those affected by genotype and ACTH. Because dysregulation of glucocorticoid production results in diverse diseases, elucidation of the function of these genes in adrenals will lead to better understanding the molecular basis of such pathological conditions.
Methods
Animals and housing
Seven-week-old male Large White (LW) and Meishan (MS) piglets (n = 24) were used in this study. The animals were reared and experiments were conducted at the INRA experimental research farm of Le Magneraud (Surgères, France). Piglets were weaned at four weeks of age and then allocated into groups of 24 animals in 3.65 × 1.70 m pens. Animals were housed in collective pens on a 10 h light, 14 h dark cycle (natural photoperiod) with food and water ad libitum. Experimental groups of six pigs of both genotypes were randomly constituted and placed in collective pens one week before the experiments. For each genotype, pigs used in this study were taken from five litters resulting from matings with two boars. The experiments described here fully comply with legislation on research involving animal subjects according to the European Communities Council Directive of November 24, 1986 (86/609/EEC). Investigators were certificated by the French governmental authority for carrying out these experiments.
Treatment and sampling
Piglets were either non-treated or injected in the neck muscle with mammalian 1–24 ACTH (Immediate Synachten, Novartis France) at the dose of 250 μg per animal. The dose of ACTH was chosen to ensure a maximum cortisol release [30]. Non-treated animals have been chosen as control instead of animals injected with vehicle in order to have the most accurate basal conditions. In accordance with approved slaughter methods, piglets were stunned and immediately exsanguinated after capture from their home pen (non-treated animals) or one hour following ACTH injection. Experiments were performed between 08.00 h and 10.00 h. Blood samples were collected directly from each piglet in heparined tubes at sacrifice. The blood was kept on ice until centrifugation (4000 g for 10 min) and plasma frozen at -80°C until measurement of cortisol. The adrenal glands were also collected, frozen immediately on dry ice and then stored at -80°C until RNA isolation.
Cortisol measurement
Plasma total cortisol was measured using a specific radio immunoassay (as previously described in Désautés et al [7]). The cortisol data were transformed to base 10 logarithmic scores and analyzed by ANOVA to assess the effects of genotype, treatment and their interaction. Results are given as the mean ± standard error.
Total RNA isolation and purification
For each biological sample, entirely left adrenal gland was homogenized in TRIzol reagent (Invitrogen Life Technologies) and a part of the homogenized sample was then used for total RNA isolation, followed by column purification (RNeasy MinElute kit, Qiagen). This procedure ensures to get equal proportions of cortex and medulla between samples. DNA was digested using an RNase-free DNase set (Qiagen) during RNA purification. Total RNA was quantified by spectrophotometer (NanoDrop®) and its integrity was assessed on an Agilent 2100 Bioanalyser (RNA 6000 Nano LabChip, Agilent Technologies).
Microarray analysis
Gene expression was analyzed by hybridization of non commercial nylon cDNA microarrays (accession in Gene Expression Omnibus data sets: GPL3729) developed by the Biological Resources Center GADIE (Genomic for animals of economical importance, INRA France) and consisted of PCR products from 8959 cDNA clones [53]. cDNA from luciferase was present on the array as positive control (193 spots) and water was also included as negative control (64 spots). cDNA clones came from pig normalized multi-tissues libraries including adrenal glands collected from control and stress pigs. Microarrays were first hybridized with a 33P-labeled oligonucleotide sequence common to all PCR products to control the quality of spotting and quantity of target DNA accessible in each spot. Microarrays were then hybridized with 33P-labeled complex probes synthesized and labelled from 5 μg of total RNA with Supersript II RNAse H- reverse transcriptase (Invitrogen). mRNA from luciferase were added to the pigs samples for calibration. Hybridizations were carried out during 24 hours at 68°C. After washing, arrays were exposed for six to 12 hours to radioisotopic-sensitive imaging plates. Detection scanning was done with a FUJI BAS 5000 phosphoimager (Fujifilm) at 25-μm resolution and quantification of hybridization signals with the AGScan software [54]. One microarray hybridization per animal was done giving six biological replicates per experimental point.
Microarray data normalization and statistical analysis
Before statistical analysis, data were log10 transformed and centred by median for each array and each gene. A filter procedure eliminated non informative transcripts on the basis of being well measured (expression level > mean + 2 standard deviations of background signal) in 100% of the samples. Statistical analyses were done using the R software (version 2.2.1, [55]). A linear model was used to test the effect of genotype and treatment as well as their interaction, and variation in quantity of target DNA accessible in each spot was included as covariate. False discovery rate (FDR) was determined using the Benjamini-Hochberg procedure [56].
The microarray data from this research has been deposited in the NCBI Gene Expression Omnibus data repository under accession number GSE8377[57].
Functional annotation
Transcripts significantly affected by genotype and/or ACTH treatment were annotated for their function according to Gene Ontology database [58] and Expression Analysis Systematic Explorer (EASE) software from DAVID bioinformatics database [59].
Analysis of RNA changes by relative quantitative real-time PCR
To verify changes in gene expression, real-time PCR was carried out on 22 selected genes. RNA (4 μg) was reverse transcribed in a total volume of 20 μl using 200U of Superscript II (Invitrogen) reverse transcriptase, 100 pmol oligo-dT22V, 0.5 mM deoxy-NTP, and 40U RNasin (Promega). The resultant cDNA was diluted 1:100 with nuclease-free water. Five microliters of diluted cDNA was used in subsequent PCR reactions. All primers were designed based on nucleotide sequences in Genbank using the Primer Express software (PE Applied Biosystems) (table 3). PCR reaction efficiency was calculated for each primer pair with five dilution points of the calibrator sample to validate primers. Introns-exons organisation of the porcine genes was deduced by comparison with human genes using ICCARE software and primers from one pair were chosen in distinct exons of the corresponding gene. Each real-time PCR reaction consisted of 1× Power SYBR Green PCR Master Mix (PE Applied Biosystems), 0.5 μM forward and reverse primers and cDNA to a total volume of 20 μl. Reactions were carried out on an ABI PRISM 7500 Sequence detection system (PE Applied Biosystems) for 40 cycles (95°C for 15 s, 60°C for 1 min). The fold change in expression of each gene was calculated using the ΔΔCt method with the levels of transketolase RNA as an internal control; as determined by quantitative RT-PCR, the levels of transketolase did not change depending on genotype or treatment in our study (data not shown) and transketolase gene has previously been used to normalize data from quantitative RT-PCR in adrenal cells [25]. Quantitative real-time PCR analysis was done in each out of the six animals constituting an experimental point and measurements were done in duplicate. ANOVA was conducted on relative expression to assess the effects of genotype, treatment and their interaction.
Table 3.
Gene-specific primers used for quantitative RT-PCR
| Gene symbol | Forward Primer | Reverse Primer | accession no. |
| Dab2 | CCCGTGATGTGACAGACAACC | ACTAATGGCTCTGCCTGTTGC | BX918681 |
| A2m | ACGTGAGCCGAACAGAGGTC | GCGATGGCAAACTCAGCTG | BX674240 |
| Anpep | CTCATTCGGAAGCAAGACGC | CCACCGCCATAGTCCTGAAA | BX665286 |
| Fxc1 | TCCTTCCAGGAGGCCTGTC | GCTGTACCAGGGCAGGCAT | BX674767 |
| Rnf2 | CACTGTGTTAAATGGCTCTTTTTCTT | TGTGCTCCTTTGTGGGTGC | BX673517 |
| Gadd45b | GCTGATGAATGTGGACCCTGA | CCTGACACCCGCACGATATC | BX671980 |
| Eif1b | GTTTCTCTTGGAGGTTGGCATT | TCACGAGGCAGCCAAACTG | BX926052 |
| Crem | AACACGCAAACGAGAGCTGAG | GCACAGCCACACGGTTTTC | BX670994 |
| Tkt | GGACAGGAAGCTCATTCTCGA | AGCAGCCACTGCCTCACCTA | BX925610 |
| Star | GAAGAGCTTGTGGAGCGCAT | AGCCAACTCATGGGTGATGAC | U53020 |
| Scarb1 | TGTGGTTTGCAGAGAGCGG | ATGAACAGCAGGACGCAGC | NM_213967 |
| Cyp21 | TGCTTCACCACCCTGAGATTC | GCCCAGCTCGCGATCTAAC | BX916139 |
| Cyp11a1 | AGACACTGAGACTCCACCCCA | GACGGCCACTTGTACCAATGT | BX674077 |
| Ldlr | GCCTCACAGGCTCGGACATA | ACACCAGTTCACCCCTCTCG | BX673438 |
| Sqle | TGGTCCAGTTGCGCTGATT | GGGCTCCGATTTAAAGCAAAA | BX920102 |
| Bzrp | GGCACACTCTACTCGGCCAT | ACAGCCTCCTCCGAGAAGCT | BX925849 |
| Ckb | TTCACCCGCTTCTGCAATG | AGGTCAGGATGTAGCCCAGGT | BX920566 |
| Hsd3b1 | TTCCGCCCTCTCTGAGGTACT | GGTCACGAAGTGGCGATTG | BX919321 |
| Bmal1 | TCCTAGCCAACGTCCTGGAA | TCTTTGGGCCACCTTCTCC | EF216896 |
| Cry1 | TGAACCACGCTGAGGCAAG | GGATTAGATGGCACTGACGCA | BQ600826 |
| Per2 | GACGTGCCGGAATGTGTTTAC | GCTCCCGGTTTCTGTGACTC | CF789448 |
| Mc2r | ACCATGTCCCCGCAGTGAT | GTGATGGCCCCTTTCATGTT | AF450083 |
| Cyp11b | CCCGTGGGTATCTTCTTGGA | GGTTTCGACCCAGGGAGTAGA | D38590 |
Gene-specific primers were designed based on sequences provided under the accession numbers listed in the National Center for Biotechnology Information (NCBI) database.
Abbreviations
ACTH, adrenocorticotropic hormone; Alas1, aminolevulinate delta synthase 1; Bmal1 (or Arntl), Brain and muscle ARNT-like 1 (or aryl hydrocarbon receptor nuclear translocator-like protein 1); Bzrp (or TSPO), peripheral-type benzodiazepine receptor (or translocator protein); CBG, corticosteroid-binding globulin; Crem, cAMP responsive element modulator; CRH, corticotropin-releasing hormone; Cry1, cryptochrome 1; Cyp11a1, cytochrome P450 family 11 subfamily B polypeptide 1; Cyp11b, cytochrome P450 family 11 subfamily B; Cyp17, cytochrome P450 family 17; Cyp21, cytochrome P450 family 21; ERK, extra-cellular signal regulated kinase; Fxc1, fracture callus 1 homolog (rat); Hmgcr, hydroxymethylglutaryl coenzymeA reductase; HPA, hypothalamic-pituitary-adrenal; 3βhsd (or Hsd3b1), 3β hydroxysteroid dehydrogenase; Ldlr, low density lipoprotein receptor; LW, Large White; MAPK, mitogen activating protein kinase; Mc2r, melanocortin 2 receptor; Mdh2, malate dehydrogenase 2 NAD (mitochondrial); MS, Meishan; P450scc, cytochrome P450 cholesterol side chain cleavage enzyme; Per2, period homolog 2; Rnf2, ring finger protein 2; Scarb1, scavenger receptor beta 1; Sdha, succinate dehydrogenase complex subunit A flavoprotein (Fp); Snf1lk, SNF1-like kinase; Sqle, squalene epoxidase; Star, steroidogenic acute regulator; Suclg2, succinate-CoA ligase GDP-forming beta subunit; TCA, tricarboxylic acid.
Authors' contributions
DH was involved in experimental design and in planning the study, carried out experiments on animals, performed radioimmunoassay, extracted RNA, carried out microarray molecular work and real-time PCR work, performed statistical analysis, interpreted data and drafted the paper. LL was involved in the microarray molecular work, contributed to the statistical analyses of the microarray data and to the writing of the manuscript. MSC designed and performed statistical analyses of microarray data and contributed to the writing of the manuscript. PM conceived of the study and coordinated the experimental design, carried out experiments on animals, contributed to the RNA extraction and to the microarray molecular work, to the analysis and interpretation of data, and to writing the paper. All authors have read and approved the final manuscript.
Supplementary Material
Complete list of genes differentially expressed depending on genotype and/or ACTH treatment. The data include one table.
Means and standard errors (SE) of transcripts levels (log) and Pearson correlation coefficients between real-time PCRs and microarray data. The data include one table.
Acknowledgments
Acknowledgements
The present results were obtained as part of a larger integrated research project SABRE [60] supported by funding under the 6th Research Framework Programme of the European Union. D. Hazard was supported by grants from Institut National de la Recherche Agronomique for completion of a post-doctoral contract.
We thank Dr Byron Jones for his valuable contribution to improving the quality of the manuscript. The authors also thank Yvon Billon and his team at the "Domaine Experimental du Magneraud" for expert technical assistance in pigs' experimentation. We also wish to thank the SIGENAE group that provides bioinformatics tools for the microarray study [61]. Finally, we acknowledge support from the CRGS platform of the Toulouse Midi-Pyrénées Génopôle [62] where part of microarray experiments was realized.
Contributor Information
Dominique Hazard, Email: dominique.hazard@bordeaux.inra.fr.
Laurence Liaubet, Email: laurence.liaubet@toulouse.inra.fr.
Magali SanCristobal, Email: magali.san-cristobal@toulouse.inra.fr.
Pierre Mormède, Email: pierre.mormede@bordeaux.inra.fr.
References
- McEwen BS. Physiology and neurobiology of stress and adaptation: central role of the brain. Physiol Rev. 2007;87:873–904. doi: 10.1152/physrev.00041.2006. [DOI] [PubMed] [Google Scholar]
- Kirschbaum C, Wust S, Faig HG, Hellhammer DH. Heritability of cortisol responses to human corticotropin-releasing hormone, ergometry, and psychological stress in humans. J Clin Endocrinol Metab. 1992;75:1526–1530. doi: 10.1210/jc.75.6.1526. [DOI] [PubMed] [Google Scholar]
- Jones BC, Sarrieau A, Reed CL, Azar MR, Mormède P. Contribution of sex and genetics to neuroendocrine adaptation to stress in mice. Psychoneuroendocrinology. 1998;23:505–517. doi: 10.1016/S0306-4530(98)00014-6. [DOI] [PubMed] [Google Scholar]
- Gomez F, Lahmame A, de Kloet ER, Armario A. Hypothalamic-pituitary-adrenal response to chronic stress in five inbred rat strains: differential responses are mainly located at the adrenocortical level. Neuroendocrinology. 1996;63:327–337. doi: 10.1159/000126973. [DOI] [PubMed] [Google Scholar]
- Sarrieau A, Chaouloff F, Lemaire V, Mormède P. Comparison of the neuroendocrine responses to stress in outbred, inbred and F1 hybrid rats. Life Sci. 1998;63:87–96. doi: 10.1016/S0024-3205(98)00243-4. [DOI] [PubMed] [Google Scholar]
- Sarrieau A, Mormède P. Hypothalamic-pituitary-adrenal axis activity in the inbred Brown Norway and Fischer 344 rat strains. Life Sci. 1998;62:1417–1425. doi: 10.1016/S0024-3205(98)00080-0. [DOI] [PubMed] [Google Scholar]
- Désautés C, Bidanel JP, Mormède P. Genetic study of behavioral and pituitary-adrenocortical reactivity in response to an environmental challenge in pigs. Physiology and Behavior. 1997;62:337–345. doi: 10.1016/S0031-9384(97)00019-X. [DOI] [PubMed] [Google Scholar]
- Satterlee DG, Johnson WA. Selection of Japanese quail for contrasting blood corticosterone response to immobilization. Poultry Science. 1988;67:25–32. doi: 10.3382/ps.0670025. [DOI] [PubMed] [Google Scholar]
- Edens FW, Siegel HS. Adrenal responses in high and low ACTH response lines of chickens during acute heat stress. General and Comparative Endocrinology. 1975;25:64–73. doi: 10.1016/0016-6480(75)90040-4. [DOI] [PubMed] [Google Scholar]
- Hazard D, Couty M, Richard S, Guémené D. Intensity and duration of corticosterone response to stressful situations in Japanese quail divergently selected for tonic immobility. Gen Comp Endocrinol. 2008;155:288–297. doi: 10.1016/j.ygcen.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Duclos M, Corcuff JB, Etcheverry N, Rashedi M, Tabarin A, Roger P. Abdominal obesity increases overnight cortisol excretion. J Endocrinol Invest. 1999;22:465–471. doi: 10.1007/BF03343591. [DOI] [PubMed] [Google Scholar]
- Pasquali R, Vicennati V, Gambineri A. Adrenal and gonadal function in obesity. J Endocrinol Invest. 2002;25:893–898. doi: 10.1007/BF03344053. [DOI] [PubMed] [Google Scholar]
- Foury A, Geverink NA, Gil M, Gispert M, Hortos M, Font i Furnols M, Carrion D, Blott SC, Plastow GS, Mormède P. Stress neuroendocrine profiles in five pig breeding lines and the relationship with carcass composition. Animal. 2007;1:973–982. doi: 10.1017/S1751731107000249. [DOI] [PubMed] [Google Scholar]
- Désautés C, Sarrieau A, Caritez JC, Mormède P. Behavior and pituitary-adrenal function in large white and Meishan pigs. Domestic Animal Endocrinology. 1999;16:193–205. doi: 10.1016/S0739-7240(99)00014-4. [DOI] [PubMed] [Google Scholar]
- Mormède P, Courvoisier H, Ramos A, Marissal-Arvy N, Ousova O, Désautés C, Duclos M, Chaouloff F, Moisan MP. Molecular genetic approaches to investigate individual variations in behavioral and neuroendocrine stress responses. Psychoneuroendocrinology. 2002;27:563–583. doi: 10.1016/S0306-4530(01)00093-2. [DOI] [PubMed] [Google Scholar]
- Bertagna X, Coste J, Raux-Demay MC, Letrait M, Strauch G. The combined corticotropin-releasing hormone/lysine vasopressin test discloses a corticotroph phenotype. J Clin Endocrinol Metab. 1994;79:390–394. doi: 10.1210/jc.79.2.390. [DOI] [PubMed] [Google Scholar]
- Moncek F, Kvetnansky R, Jezova D. Differential responses to stress stimuli of Lewis and Fischer rats at the pituitary and adrenocortical level. Endocr Regul. 2001;35:35–41. [PubMed] [Google Scholar]
- Zhang SH, Hennessy DP, Cranwell PD. Pituitary and adrenocortical responses to corticotropin-releasing factor in pigs. Am J Vet Res. 1990;51:1021–1025. [PubMed] [Google Scholar]
- Zhang SH, Hennessy DP, McCauley I, Cranwell PD. Adrenocortical ACTH receptors in pigs of differing in vivo response to adrenocorticotropin. Comparative Biochemistry and Physiology Part A, Physiology. 1993;104:43–49. doi: 10.1016/0300-9629(93)90006-P. [DOI] [PubMed] [Google Scholar]
- Ousova O, Guyonnet-Duperat V, Iannuccelli N, Bidanel JP, Milan D, Genet C, Llamas B, Yerle M, Gellin J, Chardon P, Emptoz-Bonneton A, Pugeat M, Mormède P, Moisan MP. Corticosteroid binding globulin: a new target for cortisol-driven obesity. Mol Endocrinol. 2004;18:1687–1696. doi: 10.1210/me.2004-0005. [DOI] [PubMed] [Google Scholar]
- Stocco DM, Wang XJ, Jo Y, Manna PR. Multiple signaling pathways regulating steroidogenesis and steroidogenic acute regulatory protein expression: more complicated than we thought. Mol Endocrinol. 2005;19:2647–2659. doi: 10.1210/me.2004-0532. [DOI] [PubMed] [Google Scholar]
- Sewer MB, Waterman MR. ACTH modulation of transcription factors responsible for steroid hydroxylase gene expression in the adrenal cortex. Microsc Res Tech. 2003;61:300–307. doi: 10.1002/jemt.10339. [DOI] [PubMed] [Google Scholar]
- Stocco DM, Clark BJ. Regulation of the acute production of steroids in steroidogenic cells. Endocr Rev. 1996;17:221–244. doi: 10.1210/er.17.3.221. [DOI] [PubMed] [Google Scholar]
- Simpson ER, Waterman MR. Regulation of the synthesis of steroidogenic enzymes in adrenal cortical cells by ACTH. Annu Rev Physiol. 1988;50:427–440. doi: 10.1146/annurev.ph.50.030188.002235. [DOI] [PubMed] [Google Scholar]
- Schimmer BP, Cordova M, Cheng H, Tsao A, Goryachev AB, Schimmer AD, Morris Q. Global profiles of gene expression induced by adrenocorticotropin in Y1 mouse adrenal cells. Endocrinology. 2006;147:2357–2367. doi: 10.1210/en.2005-1526. [DOI] [PubMed] [Google Scholar]
- Schimmer BP, Cordova M, Cheng H, Tsao A, Morris Q. A genome-wide assessment of adrenocorticotropin action in the Y1 mouse adrenal tumor cell line. Molecular and Cellular Endocrinology. 2007;265-266:102–107. doi: 10.1016/j.mce.2006.12.024. [DOI] [PubMed] [Google Scholar]
- Lee JJ, Widmaier EP. Gene array analysis of the effects of chronic adrenocorticotropic hormone in vivo on immature rat adrenal glands. Journal of Steroid Biochemistry and Molecular Biology. 2005;96:31–44. doi: 10.1016/j.jsbmb.2005.01.026. [DOI] [PubMed] [Google Scholar]
- Clark BJ, Wells J, King SR, Stocco DM. The purification, cloning, and expression of a novel luteinizing hormone-induced mitochondrial protein in MA-10 mouse Leydig tumor cells. Characterization of the steroidogenic acute regulatory protein (StAR) J Biol Chem. 1994;269:28314–28322. [PubMed] [Google Scholar]
- Lehoux JG, Fleury A, Ducharme L. The acute and chronic effects of adrenocorticotropin on the levels of messenger ribonucleic acid and protein of steroidogenic enzymes in rat adrenal in vivo. Endocrinology. 1998;139:3913–3922. doi: 10.1210/en.139.9.3913. [DOI] [PubMed] [Google Scholar]
- Hennessy DP, Stelmasiak T, Johnston NE, Jackson PN, Outch KH. Consistent capacity for adrenocortical response to ACTH administration in pigs. Am J Vet Res. 1988;49:1276–1283. [PubMed] [Google Scholar]
- Yao B, Rakhade SN, Li Q, Ahmed S, Krauss R, Draghici S, Loeb JA. Accuracy of cDNA microarray methods to detect small gene expression changes induced by neuregulin on breast epithelial cells. BMC Bioinformatics. 2004;5:99. doi: 10.1186/1471-2105-5-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, He YD, Kidd MJ, King AM, Meyer MR, Slade D, Lum PY, Stepaniants SB, Shoemaker DD, Gachotte D, Chakraburtty K, Simon J, Bard M, Friend SH. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/S0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- Le Roy C, Li JY, Stocco DM, Langlois D, Saez JM. Regulation by Adrenocorticotropin (ACTH), Angiotensin II, Transforming Growth Factor-{beta}, and Insulin-Like Growth Factor I of bovine adrenal cell steroidogenic capacity and expression of ACTH Receptor, Steroidogenic Acute Regulatory Protein, Cytochrome P450c17, and 3{beta}-Hydroxysteroid Dehydrogenase. Endocrinology. 2000;141:1599–1607. doi: 10.1210/en.141.5.1599. [DOI] [PubMed] [Google Scholar]
- McCammon MT, Epstein CB, Przybyla-Zawislak B, McAlister-Henn L, Butow RA. Global transcription analysis of Krebs tricarboxylic acid cycle mutants reveals an alternating pattern of gene expression and effects on hypoxic and oxidative genes. Mol Biol Cell. 2003;14:958–972. doi: 10.1091/mbc.E02-07-0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragan CA, Zearo S, Hannemann F, Bernhardt R, Bureik M. Efficient conversion of 11-deoxycortisol to cortisol (hydrocortisone) by recombinant fission yeast Schizosaccharomyces pombe. FEMS Yeast Res. 2005;5:621–625. doi: 10.1016/j.femsyr.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Dragan CA, Blank LM, Bureik M. Increased TCA cycle activity and reduced oxygen consumption during cytochrome P450-dependent biotransformation in fission yeast. Yeast. 2006;23:779–794. doi: 10.1002/yea.1383. [DOI] [PubMed] [Google Scholar]
- De Matteis F. Toxicological aspects of liver heme biosynthesis. Semin Hematol. 1988;25:321–329. [PubMed] [Google Scholar]
- Martini CN, de Avalos SGV, Romero DG, de Viale LSM, del Carmen Vila M. Heme availability affects corticosterone and aldosterone biosynthesis in rat adrenal. Steroids. 1997;62:767–770. doi: 10.1016/S0039-128X(97)00074-3. [DOI] [PubMed] [Google Scholar]
- Grion N, Repetto EM, Pomeraniec Y, Calejman CM, Astort F, Sanchez R, Pignataro OP, Arias P, Cymeryng CB. Induction of nitric oxide synthase and heme oxygenase activities by endotoxin in the rat adrenal cortex: involvement of both signaling systems in the modulation of ACTH-dependent steroid production. J Endocrinol. 2007;194:11–20. doi: 10.1677/JOE-06-0199. [DOI] [PubMed] [Google Scholar]
- Sewer MB, Waterman MR. Adrenocorticotropin/cyclic adenosine 3',5'-monophosphate-mediated transcription of the human CYP17 gene in the adrenal cortex is dependent on phosphatase activity. Endocrinology. 2002;143:1769–1777. doi: 10.1210/en.143.5.1769. [DOI] [PubMed] [Google Scholar]
- Mountjoy KG, Bird IM, Rainey WE, Cone RD. ACTH induces up-regulation of ACTH receptor mRNA in mouse and human adrenocortical cell lines. Mol Cell Endocrinol. 1994;99:R17–20. doi: 10.1016/0303-7207(94)90160-0. [DOI] [PubMed] [Google Scholar]
- Carey LC, Su Y, Valego NK, Rose JC. Infusion of ACTH stimulates expression of adrenal ACTH receptor and steroidogenic acute regulatory protein mRNA in fetal sheep. Am J Physiol Endocrinol Metab. 2006;291:E214–220. doi: 10.1152/ajpendo.00578.2005. [DOI] [PubMed] [Google Scholar]
- Winnay JN, Hammer GD. Adrenocorticotropic Hormone-Mediated Signaling Cascades Coordinate a Cyclic Pattern of Steroidogenic Factor 1-Dependent Transcriptional Activation. Mol Endocrinol. 2006;20:147–166. doi: 10.1210/me.2005-0215. [DOI] [PubMed] [Google Scholar]
- Manna PR, Dyson MT, Eubank DW, Clark BJ, Lalli E, Sassone-Corsi P, Zeleznik AJ, Stocco DM. Regulation of steroidogenesis and the Steroidogenic Acute Regulatory Protein by a member of the cAMP Response-Element Binding Protein Family. Mol Endocrinol. 2002;16:184–199. doi: 10.1210/me.16.1.184. [DOI] [PubMed] [Google Scholar]
- Kraemer FB. Adrenal cholesterol utilization. Molecular and Cellular Endocrinology. 2007;265-266:42–45. doi: 10.1016/j.mce.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Papadopoulos V, Liu J, Culty M. Is there a mitochondrial signaling complex facilitating cholesterol import? Molecular and Cellular Endocrinology. 2007;265-266:59–64. doi: 10.1016/j.mce.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Miller WL, Strauss JF. Molecular pathology and mechanism of action of the steroidogenic acute regulatory protein, StAR. Journal of Steroid Biochemistry and Molecular Biology. 1999;69:131–141. doi: 10.1016/S0960-0760(98)00153-8. [DOI] [PubMed] [Google Scholar]
- Liu J, Heikkila P, Meng QH, Kahri AI, Tikkanen MJ, Voutilainen R. Expression of low and high density lipoprotein receptor genes in human adrenals. Eur J Endocrinol. 2000;142:677–682. doi: 10.1530/eje.0.1420677. [DOI] [PubMed] [Google Scholar]
- Oster H, Damerow S, Hut RA, Eichele G. Transcriptional profiling in the adrenal gland reveals circadian regulation of hormone biosynthesis genes and nucleosome assembly genes. J Biol Rhythms. 2006;21:350–361. doi: 10.1177/0748730406293053. [DOI] [PubMed] [Google Scholar]
- Oster H, Damerow S, Kiessling S, Jakubcakova V, Abraham D, Tian J, Hoffmann MW, Eichele G. The circadian rhythm of glucocorticoids is regulated by a gating mechanism residing in the adrenal cortical clock. Cell Metab. 2006;4:163–173. doi: 10.1016/j.cmet.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Lemos DR, Downs JL, Urbanski HF. Twenty-four-hour rhythmic gene expression in the rhesus macaque adrenal gland. Mol Endocrinol. 2006;20:1164–1176. doi: 10.1210/me.2005-0361. [DOI] [PubMed] [Google Scholar]
- Katoh Y, Takemori H, Horike N, Doi J, Muraoka M, Min L, Okamoto M. Salt-inducible kinase (SIK) isoforms: their involvement in steroidogenesis and adipogenesis. Molecular and Cellular Endocrinology. 2004;217:109–112. doi: 10.1016/j.mce.2003.10.016. [DOI] [PubMed] [Google Scholar]
- Bonnet A, Iannuccelli E, Hugot K, Benne F, Bonaldo MF, Soares MB, Hatey F, Tosser-Klopp G. A pig multi-tissue normalised cDNA library : large-scale sequencing, cluster analysis and 9K micro-array resource generation. BMC Genomics. 2008;9:17. doi: 10.1186/1471-2164-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cathelin R, Lopez F, Klopp C. AGScan: a pluggable microarray image quantification software based on the ImageJ library. Bioinformatics. 2007;23:247–248. doi: 10.1093/bioinformatics/btl564. [DOI] [PubMed] [Google Scholar]
- R language and environment for statistical computing http://www.cran.r-project.org
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Serie B. 1995;57:289–300. [Google Scholar]
- The microarray data base http://www.ncbi.nlm.nih.gov/geo/
- Gene Ontology database AmiGO http://amigo.geneontology.org/cgi-bin/amigo/go.cgi
- DAVID bioinformatics database http://david.abcc.ncifcrf.gov/ease/ease.jsp
- SABRE http://www.sabre-eu.eu/
- SIGENAE http://www.sigenae.org/
- Génopôle Toulouse Midi-Pyrénées http://genopole-toulouse.prd.fr
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Complete list of genes differentially expressed depending on genotype and/or ACTH treatment. The data include one table.
Means and standard errors (SE) of transcripts levels (log) and Pearson correlation coefficients between real-time PCRs and microarray data. The data include one table.