Figure 1. KChIP2d sequence and cardiac expression.
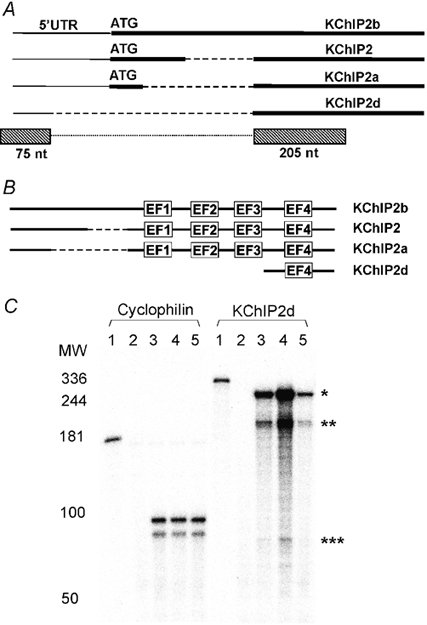
A, schematic diagram (not to scale) of KChIP nucleotide sequences. Thick lines represent coding sequence, thin lines 5′ untranslated region and dashed lines omitted regions. ATG start codons are marked. KChIP2d RNase protection assay probes represented by boxes below the schematic diagram. The dotted line connecting the boxes indicates that sequence regions in the boxes are part of the same probe without a gap in between. B, schematic diagram (not to scale) of KChIP amino acid sequences. Dashed lines represent omitted regions. EF-hands are indicated. C, RNase protection assay hybridizations of cyclophilin and KChIP2d probes (full length probes, lane 1) to yeast RNA (lane 2), ferret heart right ventricular RNA (lane 3), left ventricular epicardial RNA (lane 4) and left ventricular endocardial RNA (lane 5). Five micrograms of ferret heart RNA were hybridized to the cyclophilin control probe and 10 μg to the KChIP2d probe. Double bands in cyclophilin control hybridizations are accounted for by mismatches in the sequence of the probe (Patel et al. 2002). KChIP2d probe protected fragments: KChIP2d (*280 nt); KChIP2+2a+2b (** 205 nt and *** 75 nt). The gradient of KChIP2d expression across the ventricles parallels the distribution of the cardiac transient outward current (Näbauer et al. 1996; Brahmajothi et al. 1999).
