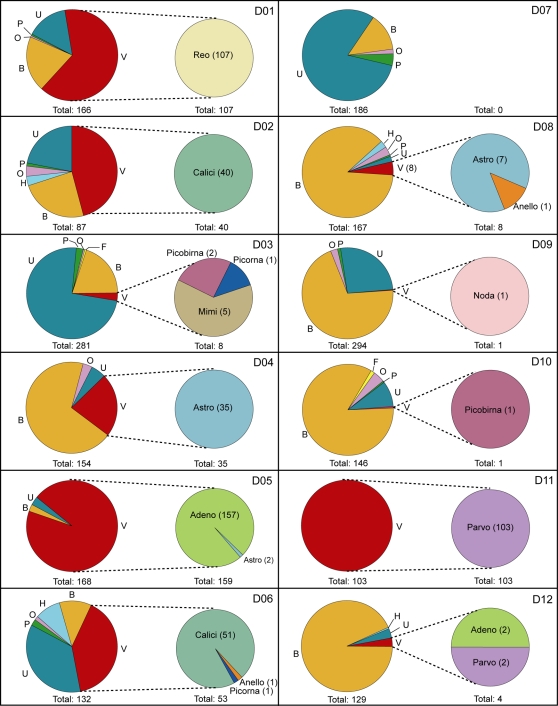
Figure 1. Categorization of sequence reads based on best tBLASTX scores (E-value: <10−5).
Pies on the left side of each box depict the categorization of sequences from individual samples by phylotype: viral (V); phage (P); bacterial (B); human (H); fungal (F); other (O); and unassigned (U). Pies on the right side of each box depict further characterization of viral sequences by viral families/taxa: Reoviridae (Reo); Caliciviridae (Calici); Astroviridae (Astro); anellovirus (Anello); picobirnavirus (Picobirna); Picornaviridae (Picorna); mimivirus (Mimi); Nodaviridae (Noda); Adenoviridae (Adeno); Parvoviridae (Parvo). Numbers in parentheses indicate the number of sequence reads in each category.