Fig. 3.
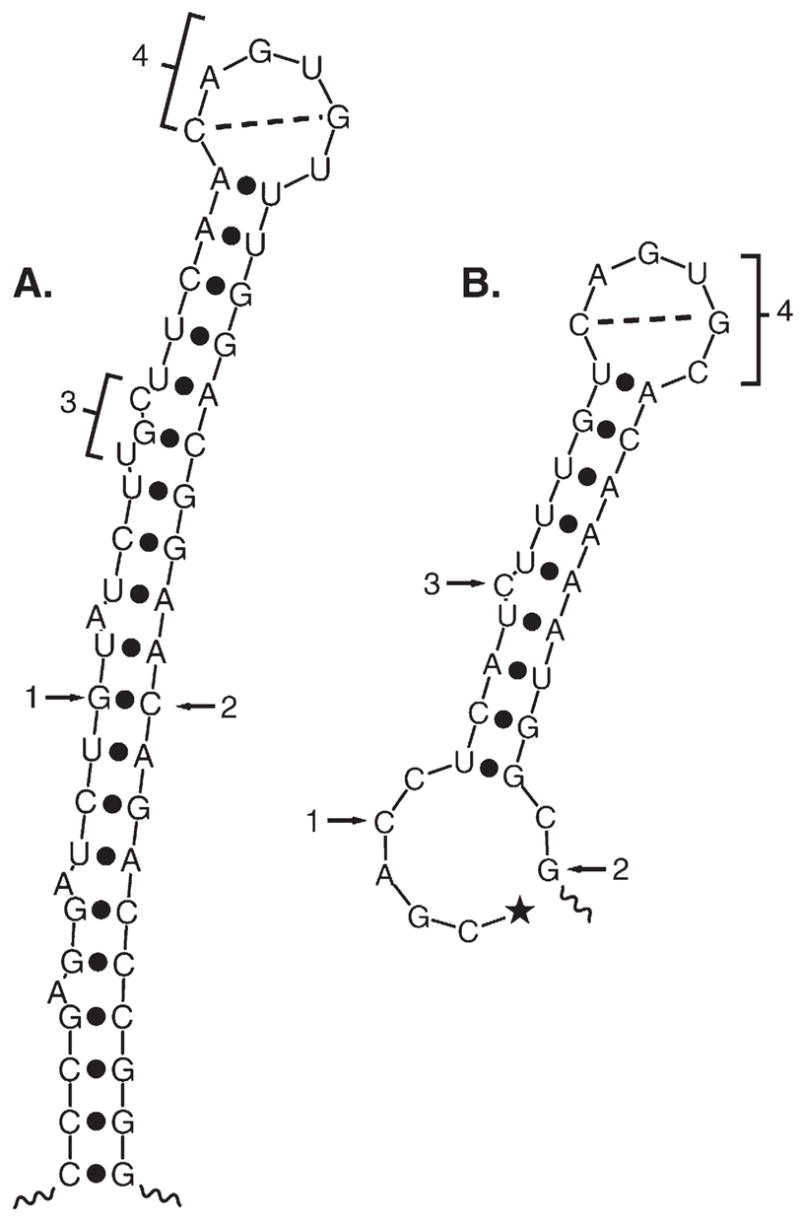
Predicted structure of ferritin and m-acon IRE regions. (A) The rat L-ferritin IRE plus flanking sequences (IRE region). Note that ferritin IRE is the central part of a large stem loop structure stabilized by sequences flanking the IRE. This structure has been confirmed in solution [143,150]. The 5′ and 3′ ends of the ferritin 28 nt IRE are indicated by arrows 1 and 2. The internal loop/bulge is indicated (bracket 3) as is the terminal loop (bracket 4). Potential basepairing between loop nucleotides 1 and 5 is shown. (B) m-acon 5′ UTR structure using the best example of an IRE-like fold obtained from computer predictions. In contrast to the ferritin and Fpn IRE regions, the m-acon IRE is not predicted to form as frequently. The highly conserved RNA sequence flanking [134] the m-acon IRE lacks extensive basepairing since, in contrast to ferritin and Fpn, the m-acon (and eALAS) IRE is <10 nt from the 5′ end. The 5′ CAP is shown for m-acon (★); ferritin cap is 28 nt from 5′ end of IRE. Structures were predicted by M-fold version 3.2.
