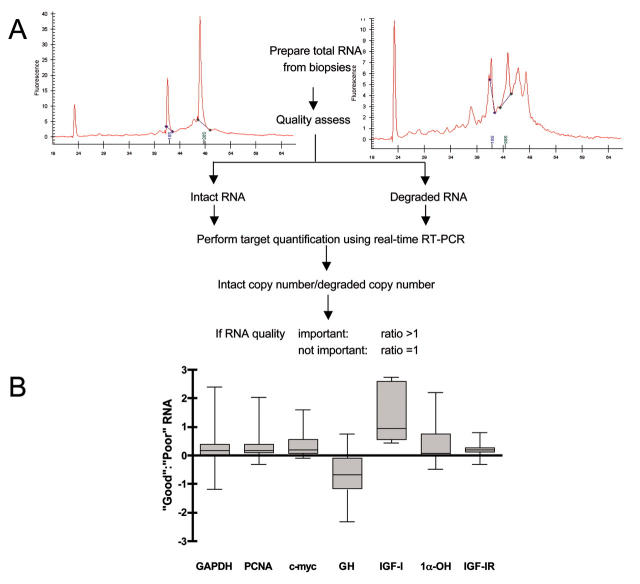
FIGURE 2.
Importance of assaying high-quality RNA. A: RNA was extracted from 19 fresh colonic biopsies using Qiagen RNeasy columns (Crawley, UK) and analyzed on the Agilent Bioanalyzer using the RNA LabChip. The intact RNA preparation on the left shows the 18S and 28S rRNA peaks as well as a small amount of 5S RNA. Degradation of the RNA sample on the right produces a shift in the RNA size distribution toward smaller fragments and a decrease in fluorescence signal as dye intercalation sites are destroyed. Where such analysis revealed degradation, RNA was re-extracted from the same sample. Real-time RT-PCR assays were carried out for seven target genes and for each sample the copy number obtained from intact RNA was divided by the copy number obtained from the degraded RNA. If RNA quality was irrelevant, the ratio of the two would be expected to be close to 1, since these are identical samples. If RNA quality was important, the ratio should be greater than 1, since the copy number calculated from the intact RNA should be higher than the copy number from the degraded RNA. B: The result shows clearly that RNA quality does matter, for some genes (e.g., IGF-I) more than for others (IGF-IR). The anomalous result obtained for GH may be due to the secondary structure of its mRNA, which is reduced by degradation, thus making it more accessible to priming. GAPDH, Glyceraldehyde-3-phosphate dehydrogenase; PCNA, proliferating cell nuclear antigen; GH, growth hormone; IGF-1, insulin-like growth factor-1; 1α-OH, 1α-hydroxylase; IGF-1R, insulin-like growth factor-1receptor.