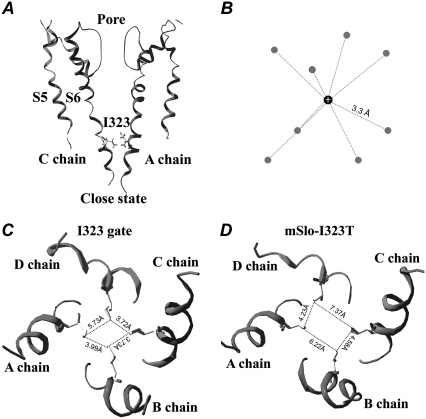
FIGURE 9.
The gating mechanism of the I323 residue in mSlo channels. (A) A closed-state structure of mSlo channels is derived from the 8-ns MD simulations. (B) A cartoon of a hydrated K+ ion is referred from the determined potassium channel structure (44). The average distance between the K+ ion and the oxygen (O) atom of a water molecule is 3.3 Å. The gray ball represents the oxygen (O) atom of a water molecule, and the black ball represents the K+ ion. (C) A regular hydrophobic ring in the mSlo channel is formed by the four I323 residues through strongly hydrophobic interactions. The distances between the neighboring I323 residues are 3.72 Å, 3.73 Å, 3.99 Å, and 5.73 Å, respectively. (D) A closed-state structure of I323T is derived from the 8-ns MD simulations; its shape exhibits a larger and more irregular rectangle than that of mSlo. The distances between the neighboring T323 residues are 4.23 Å, 7.37 Å, 6.22 Å, and 4.58 Å, respectively.