Figure 5.
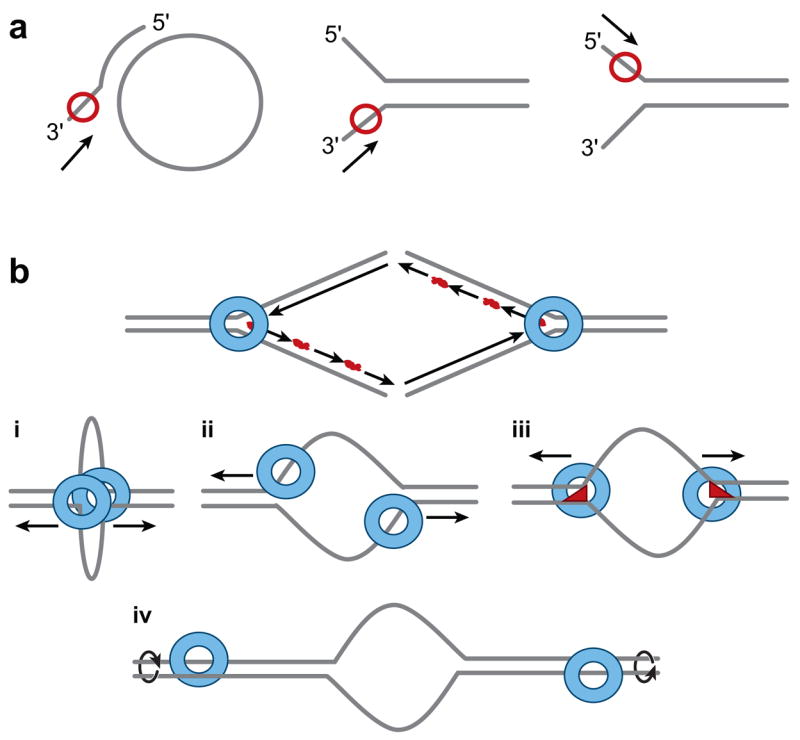
Helicase substrates and models. (a) In vitro helicase substrates that are used frequently have small ssDNA (50 bp) annealed to ssDNA circle (5 kb) with nonhomologous 3′ tail. Helicases (red circle) such as SV40 T antigen or Mcm4/6/7 complex translocate 3′ to 5′ on the tail to unwind DNA and release oligonucleotide from the larger circle. Other substrates used resemble replication forks that are produced by annealing small ssDNA oligonucleotides with nonhomologous ends. Helicases can translocate 3′ to 5′ as above or 5′ to 3′ (DnaB). (b) A single hexameric helicase is depicted as a ring (blue) at the ends of a conventionally drawn replication fork. Lagging strand Okazaki fragments are shown with RNA (red) primers at their 5′ ends. (i) The SV40 T antigen model (161) is made by putting the two rings together forming a loop. In this model, the DNA is pumped into the channel of the double hexamer and then extruded out the holes in the outside C-terminal domains (Figure 2d). (ii) In the “pump-in-ring” model, each single hexamer translocates on a different strand of DNA (127). (iii) In the “ploughshare” model, the ploughshare (red) acts as a wedge and keeps the ssDNA unwound as it emerges from behind each single hexamer (271). (iv) In the “rotary pump” model, different single hexamers twist the DNA at a distance resulting in topological strain and unwinding in the center (151).
