Abstract
A divergently oriented ampR has been located upstream of blaL2 in Stenotrophomonas maltophilia. AmpR is necessary for L1 and L2 β-lactamase induction in response to β-lactam challenge, and activation of AmpR is sufficient to induce L1 and L2 production. L1 induction requires more activation of AmpR than does L2 induction.
Stenotrophomonas maltophilia causes an increasing number of nosocomial respiratory tract and bloodstream infections in immune-compromised or severely debilitated patients. In addition, it can cause soft-tissue infections and is a frequently encountered colonizer of the lungs of cystic fibrosis patients. The majority of S. maltophilia clinical isolates are intrinsically resistant to many antimicrobials that are routinely used in hospitals, particularly β-lactams and aminoglycosides (9).
Intrinsic β-lactam resistance in S. maltophilia is due to two β-lactamase enzymes: L1, a metallo-β-lactamase with a broad substrate spectrum including carbapenems, and L2, a molecular class A, functional class 2e β-lactamase (6). In most but not all clinical isolates, L1 and L2 are produced at higher levels (induced) during β-lactam challenge (7, 11). Rare isolates that are susceptible to β-lactams do not produce β-lactamases at detectible levels, even during β-lactam challenge (11). The development of high-level resistance to weak β-lactamase inducers (e.g., ceftazidime) can be seen in the laboratory. In approximately 20% of cases, this is caused by as-yet-uncharacterized mutations that lead to constitutive β-lactamase overproduction; in the rest, there is no evidence for β-lactamase overproduction at all, and so some other, perhaps permeability- or efflux-mediated mechanism must be involved (7, 11).
The fact that both L1 and L2 are induced in response to β-lactam challenge originally led to the conclusion that a single regulatory system controls production of both enzymes. However, there is published evidence from a number of sources that mutants can be selected that overexpress only L1 or L2 whereas the partner enzyme remains inducible (1, 7). The fact that this is the case has been taken as evidence that there are two β-lactamase regulatory mechanisms in this species: one for L1 and one for L2.
In all characterized examples of β-lactamase induction systems in gram-negative bacteria, the gene encoding the transcriptional regulator is encoded immediately upstream of one of the β-lactamase genes whose expression is being regulated (13, 17). Recently, the S. maltophilia K279a genome sequence was determined. K279a is a typical clinical S. maltophilia isolate with inducible L1 and L2 (5-7, 10-12, 18). In order to investigate the mechanism by which L1 and L2 production is controlled in S. maltophilia, we started by looking upstream of blaL1 and blaL2 on the K279a genome sequence (www.sanger.ac.uk/Projects/S_maltophilia) for potential transcriptional regulator genes.
No putative upstream open reading frames were discovered that were in close proximity to blaL1. However, upstream of blaL2, a single open reading frame that has been predicted to encode a classical AmpR type β-lactamase regulator was located. The ampR-blaL2 intergenic sequence is 175 nucleotides. S. maltophilia AmpR was compared with all other AmpRs on the relevant databases using BLASTp (2), revealing that it is most similar (71% identical) to the Xanthomonas campestris AmpR regulator of its L2-like β-lactamase (19), which is perhaps not surprising given the relatively close pylogenetic relationship between these two species: S. maltophilia was once known as Xanthomonas maltophilia (9).
In order to determine whether AmpR is essential for L2 (and perhaps L1) β-lactamase induction in S. maltophilia K279a, ampR was disrupted using a suicide gene replacement vector, pEX18Tc (14), to introduce a frameshift mutation by use of the same general method described previously to disrupt K279a genes (12, 18). To do this, ampR was PCR amplified in two nonoverlapping portions by use of two primer sets. The 5′-proximal fragment had a HindIII site at its 3′ end by use of the primer, and the 3′-proximal fragment had a HindIII site introduced in its 5′ end. The primers used (with the HindIII sites underlined) were as follows: AmpRDel F (5′-CCATTCGTCGCTGCGGTAGG-3′) with AmpRHind R (5′-AAGCTTCATCCGCCTGTTCCATCGC-3′) and AmpRHind F (5′-AAGCTTTCAGTGGCAGGGT-3′) with AmpRDel R (5′-CGTCTGGCGATGACCGATG-3′). When the two resulting PCR amplicons were treated with HindIII and ligated together, the result was a mutant ampR sequence having a 121-bp deletion (and so a frameshift). The allele was named ampRFS and was ligated into pEX18Tc and mobilized into S. maltophilia K279a, and its replacement of wild-type ampR on the K279a chromosome was confirmed as set out previously (12, 14, 18).
S. maltophilia K279a and its ampRFS derivative were both subjected to imipenem challenge (10 mg/liter added at an optical density at 600 nm of 0.4 to a nutrient broth culture, which was then incubated for 2 h at 37°C with shaking). Cell extracts were prepared and specific activities of L1 and L2 β-lactamases present in these cell extracts were determined using methods described previously, which have been validated using reverse transcription-PCR to confirm that they accurately report blaL1 and blaL2 transcription levels (7). These data are presented in Table 1. Unexpectedly, they revealed that an intact AmpR is essential for induction of both L1 and L2 in K279a. MICs of a number of β-lactams against K279a and the ampRFS mutant derivate were determined using the British Society for Antimicrobial Chemotherapy standard agar dilution protocol (3) and Mueller-Hinton agar (Table 2), revealing that β-lactamase inducibility is a prerequisite for high-level β-lactam resistance in S. maltophilia; in many cases, the ampRFS mutant is acutely sensitive to β-lactams. This is reminiscent of the results seen with some clinical S. maltophilia isolates, for example, isolate J675a (7), which appear sensitive to many β-lactams in vitro and are not able to induce the production of L1 or L2 (7, 11). The fact that imipenem MICs against K279a fall following disruption of ampR (Table 2) probably explains why L1 and L2 activities drop upon challenge of the ampR mutant with 10 mg/liter imipenem (Table 1)—cells walls will be lysing, causing β-lactamase to leak into the growth medium and therefore be excluded from the assay.
TABLE 1.
Effect of disrupting ampR on β-lactamase induction in S. maltophilia K279a
| Strain | Inducer added? | L1-specific activitya | L2-specific activity |
|---|---|---|---|
| Wild-type K279a | No | 0.04 | 0.07 |
| Wild-type K279a | Yesb | 0.49 | 0.46 |
| K279a ampRFS | No | 0.08 | 0.08 |
| K279a ampRFS | Yes | 0.01 | 0.01 |
Specific activity values were calculated as described previously using nitrocefin as the substrate (7). Data represent averages of the results of three separate experiments with <20% variation.
Nutrient broth-grown cultures were induced at an OD600 of 0.4 for 2 h with 10 mg/liter imipenem.
TABLE 2.
Effect of disrupting ampR on β-lactam MICs against S. maltophilia K279a
| β-Lactam | MIC (mg/liter) against:
|
|
|---|---|---|
| Wild-type K279a | K279a ampRFS | |
| Ampicillin | 2,048 | 8 |
| Piperacillin | 2,048 | 32 |
| Ceftazidime | 128 | <4 |
| Cefotaxime | 1,024 | <4 |
| Imipenem | 64 | <4 |
| Meropenem | 64 | <4 |
The observation that AmpR is essential for L1 induction was a surprise given previous reports that it is possible to isolate L1- or L2-overproducing S. maltophilia mutants that produce the partner enzyme in an apparently normally inducible way (1, 7, 11). If AmpR directly regulates the expression of blaL1 and blaL2, then it is difficult to explain how such mutants could arise in a single-step manner. In order to investigate this apparent paradox, we isolated a number of β-lactamase-overproducing mutants of wild-type K279a by use of nutrient agar containing ceftazidime at double its MIC (2 mg/liter on this medium) as the selective agent. Mutants were then checked for β-lactamase overproduction by resuspending one colony (grown in the absence of β-lactams) in 100 μl of 100 μM nitrocefin made up in 50 mM MOPS (morpholinepropanesulfonic acid; pH 7.4) containing 100 μM ZnCl2—high-level β-lactamase production was indicated by a rapid development of red coloring. This test was carried out because of previously published reports that around 80% of high-level β-lactam-resistant K279a mutants do not actually overproduce β-lactamase (7, 11). In total, of 50 colonies tested, 12 β-lactamase hyper-producing mutants were isolated.
Specific activities of L1 and L2 were measured using our previously published method (7, 11). In the absence of β-lactam challenge, 2 of 12 mutants overproduced L2 to a much greater extent than L1, while the remainder overproduced L1 and L2. Imipenem challenge of the two L2-overproducing mutants revealed that L1 is normally inducible (Table 3). Despite repeated attempts, we could not isolate L1-overexpressing mutants in which L2 is inducible by use of isolate K279a as the parent strain. One such mutant has previously been reported (7). However, upon reanalysis of this mutant by 16S rRNA gene and smeT-smeD intergenic sequencing (our standard method for phylogenetically typing S. maltophilia) (11), we found that this mutant is not in fact derived from K279a and is instead derived from S. maltophilia isolate N531, which was being studied in parallel, and must therefore have been a contaminant (data not shown). In our previous report, isolate N531 was shown to have only an inducible L1 enzyme (7). Therefore, we are confident that S. maltophilia K279a yields mutants in which either L2 is overproduced while L1 remains inducible or both L1 and L2 are overproduced in concert.
TABLE 3.
L1- and L2-specific activities in β-lactamase hyperproducing K279a mutants
| Strain | Total nitrocefin-hydrolyzing activitya | % L1 activityb | % L2 activity |
|---|---|---|---|
| K279a | 0.05 | ||
| K279a plus inducer | 1.28 | 58 | 42 |
| M1 | 0.15 | 7 | 93 |
| M1 plus inducer | 1.51 | 66 | 34 |
| M2 | 0.23 | 5 | 95 |
| M2 plus inducer | 1.49 | 61 | 39 |
| M3 | 6.30 | 59 | 41 |
| M4 | 2.69 | 64 | 36 |
| M5 | 5.14 | 58 | 42 |
| M6 | 7.14 | 66 | 34 |
| M7 | 2.31 | 70 | 30 |
| M8 | 3.77 | 69 | 31 |
| M9 | 3.50 | 60 | 40 |
| M10 | 3.31 | 53 | 47 |
| M11c | 3.61 | 67 | 33 |
| M12 | 3.40 | 61 | 39 |
Nutrient broth cultures were grown to an OD600 of 0.4, and inducer (10 mg/liter imipenem) was added where appropriate for 2 h. Alternatively, when no induction was to be attempted, cultures were grown to an OD600 of 0.6. In all cases, cell extracts were made and total specific nitrocefin-hydrolyzing activity was determined spectrophotometrically as described previously (7). Data represent averages of the results of three separate experiments with <20% variation.
Percent contributions of L1 and L2 to total nitrocefin-hydrolyzing activity in cell extracts were calculated following addition of EDTA to reach a concentration of 100 μM (which specifically inhibits L1) to the cell extracts and reassaying using an approach described previously (7).
AmpR hyper-active mutant.
The ampR gene was sequenced from all of the K279a mutants reported in Table 3, but in only one case was a mutation found. In M11, one of the L1/L2-overexpressing mutants, a G405-to-A405 change in ampR was seen. This is predicted to cause an Asp135-to-Asn135 change in AmpR. An identical mutation has been found to constitutively activate AmpR from Enterobacter cloacae (16) and P. aeruginosa (8), and so we are confident in concluding that K279a M11 has a constitutively active AmpR. This finding allows us to definitively confirm that activation of AmpR is necessary not just for induction of L1 and L2 in the presence of β-lactam challenge but that activation of AmpR, however this is achieved, causes overproduction of L1 and L2. This demonstrates that there is no requirement for the coactivation of AmpR and another separate regulator to facilitate L1 induction.
The fact that most of the β-lactamase-overproducing mutants characterized in Table 3 have wild-type ampR genes was not a surprise. In all other studies of species with an AmpR-regulated β-lactamase, this has been found to be the case. In such organisms, β-lactamase-overproducing mutations normally occur in ampD, encoding an enzyme that breaks down the AmpR-inducing ligand. Because these ampD mutations cause a loss of function, they are more likely to occur than the specific activatory ampR mutations necessary for the same phenotype (13).
We searched for homologues of the three P. aeruginosa ampD sequences (15) by use of tBlastn searches (2) of the S. maltophilia K279a genome sequence. All three bait sequences revealed the same single hit in the S. maltophilia K279a genome, though the specificity of the hits was poor. The closest match was with P. aeruginosa ampDh2, having an E value of 10−30 and 38% identity at the amino acid level. Matches with P. aeruginosa ampD and ampDh3 were very much poorer, having E values of 10−11 and 10−17, respectively. This S. maltophilia ampD homologue was sequenced in all of the K279a β-lactamase-overproducing mutants described in Table 3, and no mutations were observed. Accordingly, it is not possible to confirm or deny the involvement of an AmpD enzyme in β-lactamase induction in S. maltophilia.
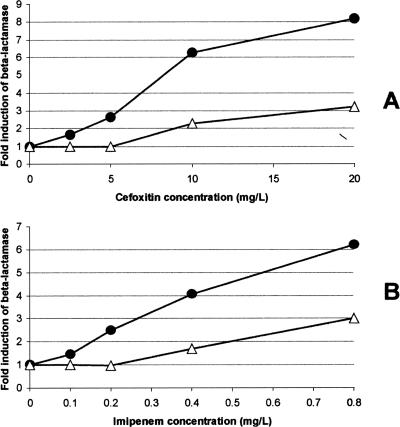
We noticed that mutants M1 and M2, which overproduce L2 significantly more than L1, did not produce as much total β-lactamase as the other mutants (including the ampR mutant, M11), which produced both L1 and L2 at high levels, and that L2 remains further inducible upon imipenem challenge, under which conditions L1 is also induced (Table 3). To explain this finding, we postulated that a greater degree of AmpR activation is necessary for induction of blaL1 expression than for induction of blaL2 expression. To test this hypothesis, we challenged wild-type K279a with increasing concentrations of imipenem or cefoxitin and specifically measured the activities of the L1 and L2 produced (Fig. 1). In both cases, production of L2 was induced at a lower concentration of β-lactam than production of L1 (Fig. 1). This adds evidence in favor of the hypothesis that while high-level activation of AmpR (by mutation or through interaction with sufficient quantities of the inducing ligand) can cause induction of L1 and L2, less activation of AmpR is required to facilitate L2 induction than to facilitate L1 induction. The molecular explanation behind this is likely to reflect either the relative binding affinities of AmpR for the blaL1 and blaL2 promoters or the involvement of an additional protein required for L1 induction which is not required for L2 induction.
FIG. 1.
Specific levels of L1 and L2 β-lactamase induction at difference inducer concentrations. Cells having an initial optical density at 600 nm of 0.4 were grown for 2 h in the presence of the inducer at the concentration stated prior to preparation of cell extracts and determination of specific activities of L1 (open triangles) and L2 (closed circles) in cell extracts as described previously (7). Briefly, total levels of nitrocefin-hydrolyzing activity (L1 and L2 activity combined) in cell extracts were determined, and the level of L2 activity alone was determined following addition of EDTA to reach a concentration of 100 μM (which specifically inhibits L1) to the cell extracts and reassaying. L1 activity was calculated by subtracting L2 activity from total activity. These specific activity values were divided by the value for the specific activity found in extracts of a culture treated identically but without addition of an inducer to give the severalfold induction values presented in the figure. Normally, we test β-lactamase inducibility in S. maltophilia by use of 100 mg/liter cefoxitin or 10 mg/liter imipenem, under which conditions each enzyme is maximally induced (as seen in Table 1). These data represent averages of the results of three experiments. There was less than 10% variation in the results obtained in these three repetitions in terms of severalfold induction levels determined for each concentration of inducer.
In conclusion, we have definitively shown that an AmpR-type regulator is necessary for the induction of L1 and L2 production in S. maltophilia. Furthermore, AmpR activation is sufficient to induce L1 and L2 production in the absence of any other stimulus. This is the first report of an AmpR regulator controlling the production of a metallo-β-lactamase and of a single AmpR positively regulating the production of more than one β-lactamase in the same cell. However, it still remains to be seen whether AmpR directly regulates L1 production (i.e., binds to the blaL1 promoter) or whether it controls the production of or (through regulating another gene) controls the activity of the direct L1 transcriptional regulator. Investigation of these possibilities will form the basis of future experimental work.
Acknowledgments
This work was funded by the Biotechnology and Biological Sciences Research Council and the British Society for Antimicrobial Chemotherapy. Sequencing of the S. maltophilia K279a genome was funded by the Wellcome Trust.
Footnotes
Published ahead of print on 22 January 2008.
REFERENCES
- 1.Akova, M., G. Bonfiglio, and D. M. Livermore. 1991. Susceptibility to β-lactam antibiotics of mutant strains of Xanthomonas maltophilia with high- and low-level constitutive expression of L1 and L2 β-lactamases. J. Med. Microbiol. 35:208-213. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Meyers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Andrews, J. M. 2001. Determining minimum inhibitory concentrations. J. Antimicrob. Chemother. 48(Suppl. 1):5-16. [DOI] [PubMed] [Google Scholar]
- 4.Avison, M. B., P. Niumsup, T. R. Walsh, and P. M. Bennett. 2000. Aeromonas hydrophila AmpH and CepH β-lactamases: derepressed expression in mutants of Escherichia coli lacking CreB. J. Antimicrob. Chemother. 46:695-702. [DOI] [PubMed] [Google Scholar]
- 5.Avison, M. B., C. J. von Heldreich, C. S. Higgins, P. M. Bennett, and T. R. Walsh. 2000. A TEM-2 β-lactamase encoded on an active Tn1-like transposon in the genome of a clinical isolate of Stenotrophomonas maltophilia. J. Antimicrob. Chemother. 46:879-884. [DOI] [PubMed] [Google Scholar]
- 6.Avison, M. B., C. S. Higgins, C. J. von Heldreich, P. M. Bennett, and T. R. Walsh. 2001. Plasmid location and molecular heterogeneity of the L1 and L2 β-lactamase genes of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 45:413-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Avison, M. B., C. S. Higgins, P. J. Ford, C. J. von Heldreich, T. R. Walsh, and P. M. Bennett. 2002. Differential regulation of L1 and L2 β-lactamase expression in Stenotrophomonas maltophilia. J. Antimicrob. Chemother. 49:387-389. [DOI] [PubMed] [Google Scholar]
- 8.Bagge, N., O. Ciofu, M. Hentzer, J. I. Campbell, M. Givskov, and N. Høiby. 2002. Constitutive high expression of chromosomal β-lactamase in Pseudomonas aeruginosa caused by a new insertion sequence (IS1669) located in ampD. Antimicrob. Agents Chemother. 46:3406-3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denton, M., and K. G. Kerr. 1998. Microbiological and clinical aspects of infection associated with Stenotrophomonas maltophilia. Clin. Microbiol. Rev. 11:57-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gould, V. C., A. Okazaki, R. A. Howe, and M. B. Avison. 2004. Analysis of sequence variation amongst smeDEF multi-drug efflux pump genes and flanking DNA from defined 16S rRNA sub-groups of clinical Stenotrophomonas maltophilia isolates. J. Antimicrob. Chemother. 54:348-353. [DOI] [PubMed] [Google Scholar]
- 11.Gould, V. C., A. Okazaki, and M. B. Avison. 2006. β-Lactam resistance and β-lactamase expression in clinical Stenotrophomonas maltophilia isolates having defined phylogenetic relationships. J. Antimicrob. Chemother. 57:199-203. [DOI] [PubMed] [Google Scholar]
- 12.Gould, V. C., and M. B. Avison. 2006. SmeDEF-mediated antimicrobial drug resistance in Stenotrophomonas maltophilia clinical isolates having defined phylogenetic relationships. J. Antimicrob. Chemother. 57:1070-1076. [DOI] [PubMed] [Google Scholar]
- 13.Hanson, N. D., and C. C. Sanders. 1999. Regulation of inducible AmpC β-lactamase expression among Enterobacteriaceae. Curr. Pharm. Des. 5:881-894. [PubMed] [Google Scholar]
- 14.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 15.Juan, C., B. Moyá, J. L. Pérez, and A. Oliver. 2006. Stepwise upregulation of the Pseudomonas aeruginosa chromosomal cephalosporinase conferring high-level β-lactam resistance involves three AmpD homologues. Antimicrob. Agents Chemother. 50:1780-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuga, A., R. Okamoto, and M. Inoue. 2000. ampR gene mutations that greatly increased AmpC β-lactamase activity in Enterobacter cloacae. Antimicrob. Agents Chemother. 44:561-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niumsup, P., A. M. Simm, K. Nurmahomed, T. R. Walsh, P. M. Bennett, and M. B. Avison. 2003. Genetic linkage of the penicillinase gene, amp, and blrAB, encoding the regulator of β-lactamase expression in Aeromonas spp. J. Antimicrob. Chemother. 51:1351-1358. [DOI] [PubMed] [Google Scholar]
- 18.Okazaki, A., and M. B. Avison. 2007. Aph(3′)-IIc, an aminoglycoside resistance determinant from Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 51:359-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weng, S. F., J. W. Lin, C. H. Chen, Y. Y. Chen, Y. H. Tseng, and Y. H. Tseng. 2004. Constitutive expression of a chromosomal class A (BJM group 2) β-lactamase in Xanthomonas campestris. Antimicrob. Agents Chemother. 48:209-215. [DOI] [PMC free article] [PubMed] [Google Scholar]