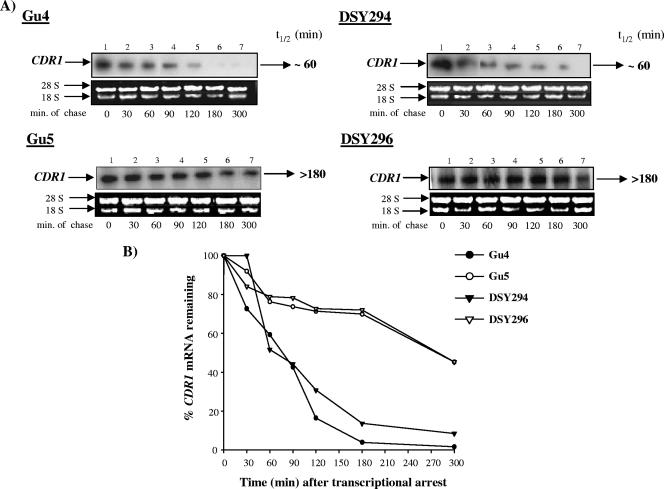
FIG. 6.
CDR1 mRNA decay assay. Exponentially growing cultures of C. albicans were incubated with the optimized thiolutin concentration (40 μg/ml) to inhibit ongoing in vivo transcription. Total RNA was isolated at the indicated times thereafter and fractionated on a 1% (wt/vol) agarose-2.2 M formaldehyde denaturing gel. (A) The gel was stained with ethidium bromide before blotting to monitor equal loading of the RNA and subsequently blotted onto a charged nylon membrane. The blot was hybridized with a CDR1-specific probe. Time points in minutes are indicated below each phosphorimage. (B) The hybridization signals were quantified using densitometry scanning in a phosphorimager. The signal intensity at each time point was normalized to that of time T0 (expressed as a percentage) and plotted as described in Materials and Methods. t1/2, half-life.