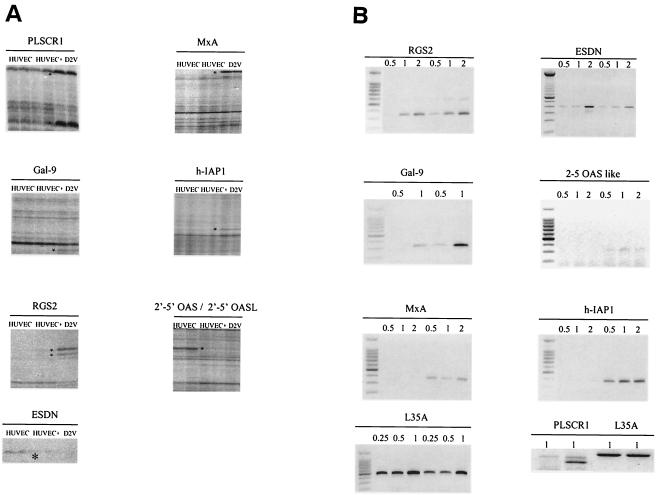
FIG. 2.
(A) Differentially displayed genes in HUVEC 48 h after D2V infection. HUVEC RNA was used to prepare cDNA with SensiScript reverse transcriptase and HT11C primer. Asterisks indicate differentially displayed bands identified in DV-infected cultures (D2V) compared to uninfected cells (HUVEC). +, addition of exogenous viral RNA to uninfected cells. (B) Semiquantitative RT-PCR analysis. One of three semiquantitative RT-PCR analyses is shown for seven of the eight genes identified by DD; uninfected (left) or virus-infected cultures (right) at 48 h postinfection are included in each panel. L35A was used as a control gene. We used 0.5, 1, and 2 μl of cDNA of either uninfected and infected samples in the PCR for all reactions except for L35A, in which 0.25, 0.5, and 1 μl of cDNA were used. The PLSCR1 gene analysis was performed independently. For the PLSCR1 RT-PCR, results obtained with 1 μl of cDNA are shown. Panel B, left side, uninfected cells; right side, infected cells. The PLSCR1 panel has uninfected cells in lanes 1 and 3 and infected cells in lanes 2 and 4.