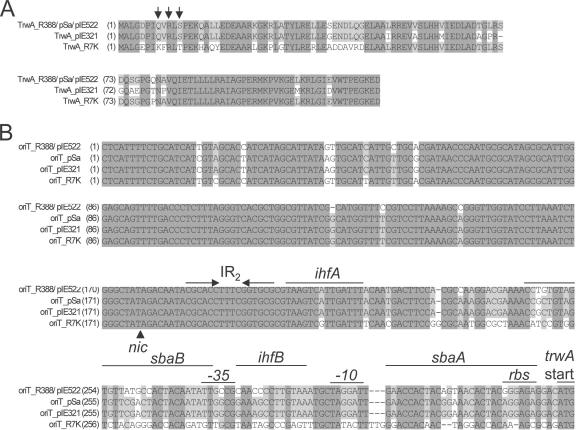
FIG. 2.
Alignments of the TrwA and oriT sequences. Alignments of the TrwA (panel A) and oriT (panel B) sequences of R388/pIE522/pSa, pIE321, and R7K are shown. Invariant nucleotides or amino acids in all plasmids are shadowed in dark gray, while those identical in most plasmids are shadowed in light gray. (A) Residues 8, 10, and 12, which are important for oriT recognition by TrwAR388, are indicated by vertical arrows. (B) The oriT DNA strand complementary to that cleaved by the relaxase is shown. The nic site is represented by a black triangle. Structural motif IR2 is represented by convergent horizontal arrows. The sites of interaction of oriTR388 with proteins IHF (ihfA and ihfB) and TrwA (sbaA and sbaB) are marked by horizontal lines. The start codon of gene trwA (trwA start), its putative ribosome binding site (rbs), and the trwA promoter −10 and −35 sequences are indicated.