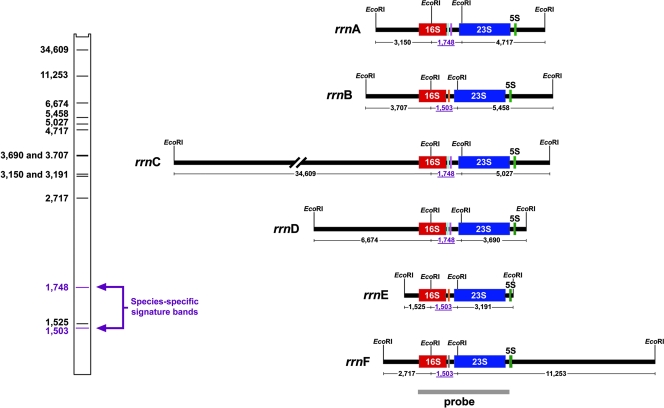
FIG. 3.
Conservation of rRNA operon sequences and variability of chromosomal flank sequences are responsible for ribotyping polymorphisms, as revealed by in silico analysis (Gene Construction Kit; Textco BioSoftware, Inc.) of the six ribosomal operons of the genomically sequenced H. influenzae strain Rd. Evolutionarily diverse isolates within a bacterial species display ribotype fragments of differing sizes comprising either the 5′ end of the 16S rRNA gene and neighboring flank DNA or the 3′ end of 23S-5S rRNA genes and neighboring flank sequence. The six ribosomal operons of strain Rd, labeled A through F, are depicted, along with EcoRI restriction sites within the rrn genes and the first flanking EcoRI site adjacent to each ribosomal operon. Based on the sizes of EcoRI fragments generated, the bar code at the left depicts the predicted EcoRI ribotype profile for strain Rd. Twelve of the 14 bands in the profile result from polymorphisms in the chromosomal flanks of the six rrn operons; the remaining two bands are the species-specific signature bands, comprising the ISR between the 16S and 23S rRNA genes and size dependent on the presence of one or two tRNA sequences. Species-specific signature band sizes are depicted beneath each operon genetic map in mauve, underlined numerals. Ribosomal operons A, C, and D contain two tRNAs (tRNAIle shown in cyan and tRNAAla shown in mauve). Ribosomal operons B, E, and F contain one tRNA (tRNAGlu shown in orange). The gray bar beneath the linear maps of the ribosomal operons depicts the location of the probe used to identify rRNA gene-containing fragments.