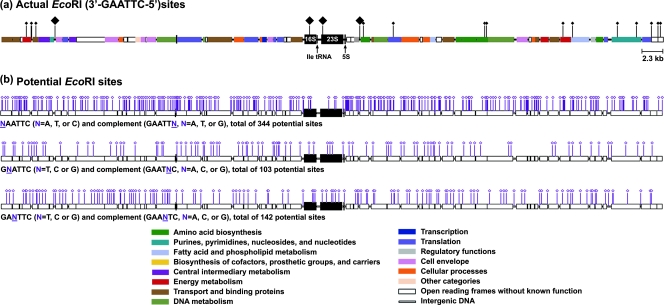
FIG. 4.
In silico analysis (Gene Construction Kit) of potential EcoRI sites in 50 kb upstream and downstream of the H. influenzae strain Rd ribosomal operon C (rrnC). Flanks of rrnC, as well as flanks of the remaining five ribosomal operons (data not shown) in H. influenzae strain Rd, are composed primarily (90.3%) of neutral housekeeping genes. Because of high intraspecies conservation of rRNA gene sequences, ribotype RFLPs found among independent H. influenzae isolates result from changes in restriction sites in the chromosomal flanks in which there exist a vast array of “potential” EcoRI sites. (a) In silico analysis of rrnC of H. influenzae strain Rd and contiguous ∼50 kb upstream and downstream sequences. Genes encoding rRNA (16S, 23S, and 5S) are depicted as large, black boxes; genes flanking the ribosomal operon are color coded according to their products’ cellular functions. Functional classification indicates that the majority of flank sites are evolutionarily neutral. Black diamonds indicate existing EcoRI restriction sites, with the four larger diamonds indicating EcoRI sites responsible for the actual RFLP profile of H. influenzae Rd. (b) In silico analysis of the ∼600 “potential” EcoRI sites that could give rise to a novel cut site by single point mutation, with each of the three maps depicting point mutations at different positions of the palindromic recognition sequence. The locations of the small diamonds reveal that these potential EcoRI sites are distributed along the two 50-kb flank sequences. EcoRI site-generating point mutations of this type in the ribosomal operon chromosomal flanks are the primary molecular genetic basis for the diversity of ribotype profiles attained. Similar in silico analysis of the remaining ribosomal operons (data not shown) allowed us to calculate the maximum possible number of ribotype RFLP profiles as 30012 = 5.3 × 1029, where 300 is the number of potential EcoRI sites for each flank and 12 is the total number of upstream and downstream flanks in any given H. influenzae isolate. As explained above in the text (see “Molecular Basis of Species Diversity Resolved by Conventional Ribotyping: Single-Point-Mutation-Based Model”), 1029 is hypothetical (i.e., the potential number of polymorphisms available by this analysis), while in reality, because of numerous biological constraints, the actual number is much smaller than the possible number.