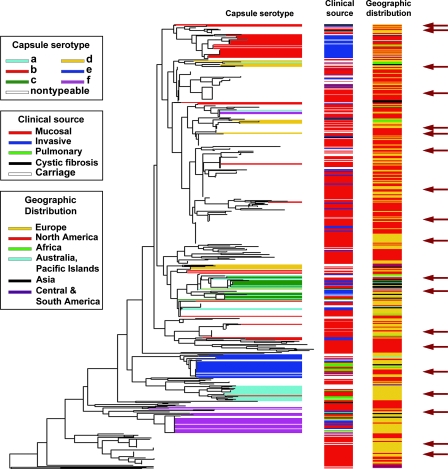
FIG. 6.
Ribotype RFLP phylogenic tree (BioNumerics; Applied Maths) based on ribotyping of ∼600 H. influenzae isolates of diverse origins. Capsular serotypes, clinical sources, and geographic distribution are depicted as horizontal barcodes. The choice of isolates to create a manageable, phylogenetically representative subset for further studies is indicated by dark arrows. Independent confirmation of this dendrogram was obtained based on capsular operon gene polymorphism analysis (44) and congruence of a ribotype-based phylogenetic tree of a subset of the type a to f isolates with results from MLEE analysis (56). Further validation was obtained by comparative gene sequencing of recA and 16S rRNA genes among a representative subset of 50 of the isolates and MLST analysis of a representative set involving 51 of the nontypeable isolates.