Abstract
In Aspergillus nidulans, proline can be used as a carbon and nitrogen source, and its metabolism requires the integration of three signals, including proline induction and nitrogen and carbon metabolite derepression. We have previously shown that the bidirectional promoter in the prnD-prnB intergenic region undergoes drastic chromatin rearrangements such that proline induction leads to the loss of positioned nucleosomes, whereas simultaneous carbon and nitrogen metabolite repression results in the partial repositioning of these nucleosomes. In the proline cluster, the inhibition of deacetylases by trichostatin A leads to partial derepression and is associated with a lack of nucleosome positioning. Here, we investigate the effect of histone acetylation in the proline cluster using strains deleted of essential components of putative A. nidulans histone acetyltransferase complexes, namely, gcnE and adaB, the orthologues of the Saccharomyces cerevisiae GCN5 and ADA2 genes, respectively. Surprisingly, GcnE and AdaB are not required for transcriptional activation and chromatin remodeling but are required for the repression of prnB and prnD and for the repositioning of nucleosomes in the divergent promoter region. Chromatin immunoprecipitation directed against histone H3 lysines K9 and K14 revealed that GcnE and AdaB participate in increasing the acetylation level of at least one nucleosome in the prnD-prnB intergenic region during activation, but these activities do not determine nucleosome positioning. Our results are consistent with a function of GcnE and AdaB in gene repression of the proline cluster, probably an indirect effect related to the function of CreA, the DNA-binding protein mediating carbon catabolite repression in A. nidulans.
In eukaryotic cells, the acetylation of histones is correlated with both transcriptional activation and the chromatin rearrangements usually associated with it. A turning point in our understanding of the mechanism of histone acetylation was the demonstration of the intrinsic acetylase activity of the Tetrahymena pyriformis p55 protein (7), the homologue of the Saccharomyces cerevisiae Gcn5p protein, which was already known to be involved in the transcriptional activation of a number of genes in this organism (reviewed in reference 33). Homologues, possibly orthologues, of Gcn5p, are universally present in eukaryotes. Gcn5p and its homologues interact with chromatin in large multiprotein complexes, such as ADA and SAGA (17). SAGA is a multiprotein complex whose Gcn5p subunit possesses histone acetyltransferase (HAT) activity. Gcn5p acetylates several lysine residues on the N termini of histones, including K9 and K14 on histone H3 and K8 and K16 on histone H4 (21). The Gcn5-containing complexes share several subunits, such as Ada2p, Ada3p, Spt3p, and Tra1p (24), and it has been estimated that the yeast SAGA complex regulates the expression of ∼10% of S. cerevisiae genes, with approximately one-third of them being negatively regulated (23). Gcn5p forms a ternary complex with Ada2p and Ada3p, a complex which is conserved in SAGA and ADA (2). The proteins of the complex modulate the acetylating activity of Gcn5p (8, 17, 35) in patterns that are not identical for different promoters or different transcriptional activators (9, 33). In the context of transcriptional regulation, the SAGA complex can have different functions. The role of SAGA has been extensively studied in the S. cerevisiae GAL1 promoter and has been shown to be essential for GAL1 transcription. In the case of GAL1, the first step in transcriptional activation is the binding of the specific Gal4p activator which, in turn, recruits SAGA to the upstream activating sequence, and the upstream activating sequence-bound SAGA then promotes the binding of TATA-binding protein and assembly of the preinitiation complex (5, 22). In contrast to the GAL1 system, in the yeast ADE regulon SAGA is not recruited by the specific activators Bas1p and Pho2p (nor is SWI/SNF, another chromatin remodeling complex recruited in this promoter). Instead, in promoters of this regulon, SAGA function is required for the recruitment and efficient binding of the specific activators (19). In the yeast nitrogen-carbon utilization regulatory interface, different components of SAGA have been shown to have different functions. The expression of both paralogous glutamate dehydrogenase genes, GDH1 and GDH3, requires different SAGA components, depending on whether glucose or ethanol is the carbon source. GDH1 expression requires Ada2p and Ada3p on ethanol as a carbon source but only Ada3p on glucose. In both cases, GDH1 expression is GCN5 independent (32). In contrast, GDH3 expression and chromatin remodeling activities in its cognate promoter, which are only seen under carbon derepressing conditions (ethanol), are dependent on the SAGA components GCN5, ADA2, ADA3, and SPT3.
There is little information on SAGA function in filamentous fungi. Recent work in Neurospora crassa established that blue light-induced transcription of the early light-inducible genes al-3 and vvd depends on increased acetylation of histone H3 at lysine K14 (H3K14) in the promoters of these genes (18). The authors showed that acetylation and transient gene activation require the N. crassa GCN5 homologue ngf-1, and it was suggested that the specific activator, White Collar-1, is required for the recruitment of NGF-1 to al-3 and vvd promoters.
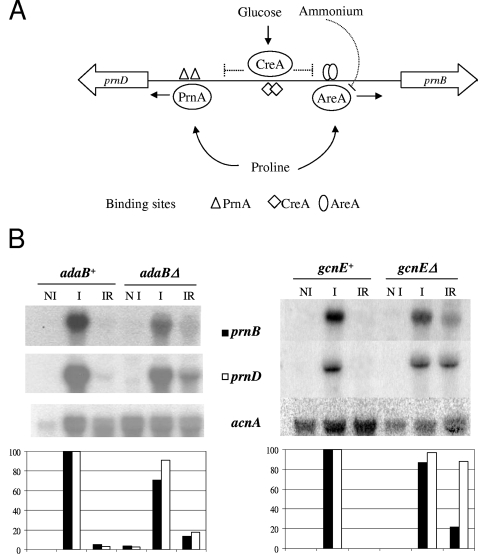
We have recently described the relationship between the transcriptional activation of a number of Aspergillus nidulans promoters driving genes of primary metabolism and their chromatin architecture (3, 12, 25, 27-29). The prn cluster of A. nidulans comprises all the genes involved in proline utilization as a sole nitrogen and/or carbon source. We have studied in detail the bidirectional prnD-prnB promoter, driving, respectively, the genes encoding proline oxidase and the specific proline transporter (10, 11, 13-15, 34). The proline metabolic genes are induced by proline and repressed when preferred carbon (glucose) or nitrogen (ammonium or glutamine) sources are available in the medium. A graphical representation of regulatory proteins and their cis-acting recognition motifs involved in the regulation of the bidirectional promoter is shown in Fig. 1A. In this promoter, eight nucleosomes lose their positioning upon induction while simultaneous carbon and nitrogen metabolite repression results in partial nucleosome repositioning. Chromatin restructuring is strictly dependent on the PrnA pathway-specific activator but not on the wide-domain nitrogen regulator AreA, which was proven to be essential for chromatin opening in the A. nidulans nitrate regulon (3, 27). AreA in the proline cluster is required only in the presence of glucose, which is consistent with its proposed function, which is to counteract the repressive effect of the global carbon repressor CreA. A functional CreA protein and two CreA binding sites in the prnD-prnB bidirectional promoter were shown to be essential for nucleosome repositioning under repression conditions (12). A first indication that histone acetylation is involved in the CreA-mediated repression of the prnD-prnB bidirectional promoter process came from the observation that trichostatin A (TSA), an inhibitor of histone deacetylases, mimics the effect of a CreA loss-of-function mutation or of cis-acting CreA binding site mutations (termed prnd). Nucleosome positioning after repression and transcriptional repression is completely lost when CreA is nonfunctional and also partially lost when TSA is present in the culture medium (12).
FIG. 1.
Regulation of proline utilization in A. nidulans. (A) Overview of proline cluster regulation. The prnD-B intergenic region is shown. prnD encodes the proline oxidase, and prnB encodes the specific proline transporter (12, 14). The pathway-specific transcription factor PrnA is essential for proline induction of both genes. In the absence of preferential carbon (glucose) and nitrogen (ammonium) sources and the presence of proline, PrnA and the GATA factor AreA bind to their cognate sites in the intergenic region, resulting in the expression of prnD and prnB. Repression requires both glucose activation of the negative regulator CreA and ammonium inactivation of AreA. Full repression occurs only in the simultaneous presence of glucose and ammonium. Repression acts directly on prnB expression; prnD repression is indirect and results from inducer exclusion. (B) Effect of adaB and gcnE deletion on prnB and prnD transcription. Strains (Table 1) were pregrown in liquid minimal medium under noninducing conditions (5 mM urea-0.1% fructose) with the appropriate supplements, harvested, divided into aliquots, and further incubated for 2 h under the conditions indicated. Noninducing (NI), 5 mM urea and 0.1% fructose; inducing (I), induced by 20 mM proline; inducing-repressing (IR), 20 mM proline and repression by 1% glucose and 20 mM ammonium-l(+)-tartrate. Expression levels (bottom) were quantified using phosphorimaging and ImageQuant software analysis. Normalized signals were obtained by comparison of specific signals with actin gene (acnA) expression signals. The induced levels in the adaB+ and gcnE+ strains are given in every case the arbitrary value of 100; filled columns represent prnB, and open columns represent prnD expression.
To investigate in more detail how acetylation affects nucleosome positioning in this system, we deleted two genes coding for members of the putative SAGA/ADA acetylation complexes in A. nidulans. In this article we describe the effect of gcnE and adaB deletions, homologues of the S. cerevisiae GCN5 and ADA2 genes, a putative histone acetylase and adaptor protein, respectively. We studied the effects of these deletions on the transcriptional competence, the chromatin structure, and the acetylation status of H3K9 and H3K14 in the prnD-prnB bidirectional promoter. Unexpectedly, low levels of histone H3 acetylation in the deletion strains were found not to affect transcriptional activation but were paradoxically associated with partial derepession of prnB and prnD.
MATERIALS AND METHODS
Strains and growth conditions.
A. nidulans strains used throughout the study are listed in Table 1. A total of 106 spores of each strain per ml were inoculated into liquid minimal medium (30) with the appropriate supplements plus 0.1% fructose as the carbon source and 5 mM urea as the nitrogen source. Mycelia were grown for 12 h at 37°C on a rotary shaker with continuous shaking at 180 rpm and then harvested by filtration. For condition-specific growth, aliquots of these precultures were then further incubated for 2 h at 37°C under the following conditions: noninducing, 0.1% fructose-5 mM urea; inducing, 20 mM proline; and inducing-repressing, 20 mM proline plus 1% glucose and 20 mM ammonium-l-(+)-tartrate. Finally, mycelia were harvested by filtration for RNA isolation, micrococcal nuclease (MNase) analysis, and chromatin immunoprecipitation (ChIP). For microscopic observations, cultures were grown on solid complete medium (30) until sporulation occurred (roughly 7 to 10 days for the adaB and gcnE deletion strains and around 3 days for the wild-type control strains), and surface samples were prepared for microscopy.
TABLE 1.
A. nidulans strains used throughout this work
| Strain | Genotype | Reference or sourcea |
|---|---|---|
| gcnE+ | pyrG89 argB2 pantoB100 riboB2 yA2 | This work |
| gcnEΔ | pyrG89 argB2 pantoB100 riboB2 yA2 gcnEΔ::pyrG+ | This work |
| adaB+ | biA1 | FGSC strain A26 |
| adaBΔ | biA1 argB::trpCΔB adaBΔ::argB+ | This work |
| MH 9233 | wA3 biA1 argB::trpCΔB pyroA4 riboB2 | 26 |
FGSC, Fungal Genetics Stock Center (http://www.fgsc.net).
Cloning of gcnE and adaB and construction of disruption vector.
Using the S. cerevisiae Gcn5p protein in a BLAST search of the A. nidulans genome database (www.broad.mit.edu), we identified gene AN3621.3 as encoding GcnE, a putative orthologue of yeast Gcn5p, with an overall identity of 66% between the A. nidulans and the S. cerevisiae proteins (for predicted domain organization and BLAST results, see http://www.broad.mit.edu/annotation/genome/aspergillus_nidulans/).
A deletion cassette was constructed by double-joint PCR. Deletion of the gcnE open reading frame (ORF) was achieved by replacing it with the A. fumigatus pyrG gene. In this cassette, the pyrG gene is flanked by upstream and downstream gcnE ORF sequences. The 3′ flanking fragment of gcnE was amplified with primers G1F (5′-TAGCAGACCCTGATGCATCAAAC-3′) and G2632R (5′-TAAAGAGCTTTCAGTAAGTAATGATACTCG-3′), and the 5′ flanking fragment of gcnE was amplified with G3330F (5′-GAAACATCCCCGAGTGGTCGGTAAGTACAA-3′) and G5950R (5′-CTCTCAAATACTAAGCCAAGTGGTACCAAG-3′) from A. nidulans wild-type (pabaA1) genomic DNA. A. fumigatus pyrG was amplified from A. fumigatus wild-type genomic DNA with primers GCNPYRF (5′-GCGTGCTACCTCTGCGAGTATCATTACTTACTGAAAGCTCTTTAGGACTGAATTCGCCTCAAACAATGC-3′) and GCNPYRR (5′-ATTGTACTTACCGACCACTGGGGATGTTTCGAATCTGCTGCCACATGAAGGAATTCTCAGTCCTGCTCC-3′). Nested primers GCNESTF (5′-GAAAATTCAACTGTCGATTCG-3′) and GCNESTR (5′-TGGGTGAAAAGTGGATTGTATG-3′) were used to amplify the complete assembled molecule containing 5′ gcnE-pyrG-3′-gcnE. This gcnE deletion cassette was used to transform a pyrG89 argB2 pantoB100 riboB2 yA2 strain. Transformants were selected on minimal medium with appropriate supplements lacking uracil and uridine.
To identify the strains with deletions of gcnE, a Southern blot with genomic DNA digested with KpnI was hybridized with a 32P-labeled probe derived from a 2.6-kb PCR product amplified with primers G3330F and G5950R. A single 3-kb band confirmed gcnE deletion and single integration of the deletion cassette. The same membrane was stripped and hybridized with a 32P-labeled probe derived from an 800-bp PCR product amplified by GCNORFF (5′-TCGTTGGAGGTATAACCTACCG-3′) and GCNORFR (5′-GTAGGGTGTGTTCTCGTTATTATACC-3′) corresponding to the gcnE ORF. No signal was detected for the strain with gcnE deleted (not shown).
Using the S. cerevisiae Ada2p protein in a BLAST search of the A. nidulans genome database (www.broad.mit.edu), we identified the product of gene AN5974.2 (changed to gene number AN10763.3 in the latest annotation) as a putative orthologue of yeast Ada2p with a similarity score of 3 × 10e−79 (for predicted domain organization and BLAST results, see http://www.broad.mit.edu/annotation/genome/aspergillus_nidulans/). The transcription start and termination points were determined empirically by 5′ and 3′ rapid amplification of cDNA ends, respectively, and were found to correspond to the sites shown in the A. nidulans database. Plasmid pMS12 (Fungal Genetics Stock Center, http://www.fgsc.net) containing the argB gene was used to insert PCR fragments derived from the adaB 5′ region upstream of argB and from the adaB 3′ region downstream of argB. The adaB 5′ and 3′ sequences were obtained by PCR amplification using the sequence information from the A. nidulans sequence database (www.broad.mit.edu). Roughly 700 bp of the adaB 5′ region was amplified with primers 5′ forward (5′-GATGCAAGACCGCGGCCGCCACAGCAGTCAGC-3′; the NotI site introduced by mutating three bases is underlined) and 5′ reverse (5′-CATCGATCCCGAGAGCCTTTCATCCTCCGCG-3′). The fragment was cloned as an NotI-ClaI fragment into the vector. Around 1.4 kb of the adaB 3′ region was amplified with primers 3′ forward (5′-CGAATCATCACGTCGACTTGAATTTAGAGGCC-3′; the SalI site introduced by mutation of the HindIII restriction site, positioned 50 bp downstream of the empirically determined polyadenylation site, is underlined) and 3′ reverse (5′-GCGAGTTGACTGAGCTCGAGAAGGATCACCCTC-3′). The fragment was cloned as an SalI-XhoI fragment into the vector. The resulting 7.2-kb vector was called p5′-3′adaB::argB. Strain MH 9233 (26) (gift from Michael Hynes, The University of Melbourne) was used as a recipient strain for the knockout construct. To verify the insertion of the argB gene at the adaB locus, Southern hybridization was carried out, and the putative deletion strains (showing the absence of the 4.5-kb adaB band and instead a single 3.5-kb band hybridizing with the 3′ adaB Hind III-XhoI probe) were verified for the absence of the adaB ORF and additional argB insertions. The strain was crossed to a wild-type pabaA1 strain to obtain an adaBΔ strain with only one additional marker for further studies. An adaBΔ biA1 strain was selected from the progeny, and for further studies, a wild-type biA1 strain was used as a reference strain.
RNA preparation and Northern blots.
Total RNA was isolated with RNA Plus extraction solution (Biogen) following the manufacturer's instructions. RNA electrophoresis and Northern blot hybridizations were carried out as described previously (13). prnB, prnD, and acnA probes were prepared as described by Gomez et al. (14).
Nucleosome positioning.
MNase I digestions were performed by the method adapted by Gonzalez and Scazzocchio (16). MNase was used at concentrations ranging from 0.5 to 2.5 U/g of mycelium. DNA was digested with an appropriate restriction enzyme. Probes SC1 and SC2 for the prnB-D intergenic region were prepared as described previously (12).
The position of each MNase cut was calculated by running in each gel a scale of molecular size markers (100 Base-Pair Ladder; Amersham Pharmacia Biotech., Piscataway, NJ). The values reported in the scale adjacent to each gel represent the position of the MNase cut from the ATG of the relevant gene. For each mutant and growth condition, the experiments were performed in triplicate.
ChIP assays.
ChIP assays were carried out following our published protocol (4). Antibodies for ChIP analysis of acetylated histone H3 were purchased from Upstate Biotechnology (Charlottesville, VA) and recognized acetylated K9 and K14 of histone H3 (06-599). Rabbit polyclonal antibody recognizing the C terminus of histone H3 (ab1791) was purchased from Abcam (Cambridge, United Kingdom). The latter antibody was used to normalize the acetylation status of H3. Two microliters of antibody solution was used to incubate 200 μg of protein.
Quantitative real-time PCR was performed using a Bio-Rad (Hercules, CA) MyiQ Cycler with the Platinum Sybr green qPCR SuperMix-UDG from Invitrogen (Karlsruhe, Germany) for amplification. Chromosomal DNA from a wild-type strain was used as an external standard for setting up the calibration curve. Primers that amplified fragment a (see Fig. 4B) had the following sequences: for prnB+2for, 5′-TGAGGATCCCATTAGTCAAGG-3′; and for prnB+2rev, 5′-GGATCAGGTTCCCTAAGATCAG-3′. The PCR was calibrated by a dilution series of total DNA extracted from a wild-type strain (3.3 μg μl−1). Each PCR was replicated (technical repetition). For each sample the absolute amount of the specific DNA fragment in the immunoprecipitated sample was divided by the amount of this fragment in the sample before precipitation (normalizing to input DNA). To obtain the relative K9K14 acetylation level of histone H3, the normalized values obtained from acetylated H3 ChIPs (H3-acetyl) were divided by the normalized values obtained from the H3 C-terminal ChIPs (H3 C-term). The H3-acetyl/H3 C-term ratio obtained in the adaB+ wild-type strain grown under noninducing conditions was set to one. Two biological repetitions were performed for each condition, and standard deviations were calculated.
RESULTS
Phenotypes of the gcnE and adaB deletions.
Both the gcnE and adaB deletion strains showed a strongly reduced growth rate and conidiation on solid medium. Both deletions resulted in similar morphological alterations of the conidiophore. Supplemental Fig. S1 shows a selection of pictures obtained from microscopic observations of the adaB deletion strain (see the supplemental material). The strain produces stalks and vesicles, but metulae are missing. Short stalks with vesicle heads and a few phialides repeatedly emerge directly from vegetative mycelia. None of the known conidiation mutants of A. nidulans displays this phenotype.
We tested for specific effects of the gcnEΔ and adaBΔ strains on the utilization of various carbon and nitrogen sources. Compared to growth of the isogenic wild-type strains, both deletion strains displayed reduced growth on all the nutrients tested, including complete medium and proline as a sole carbon or nitrogen source (data not shown).
prnD-prnB expression is partially derepressed in the gcnEΔ and adaBΔ strains.
Garcia and coworkers (12) have shown that the expression of proline catabolic genes and chromatin rearrangements in the prnD-prnB intergenic region are affected by TSA-mediated inhibition of deacetylases. Thus, we investigated the expression of prnB and prnD in the gcnEΔ and adaBΔ backgrounds. Figure 1B shows Northern blots from isogenic gcnE+ and gcnEΔ strains (right panel), as well as isogenic adaB+ and adaBΔ strains (left panel) grown under noninducing (fructose and urea), inducing (proline), and inducing-repressing (proline, glucose, and ammonium) conditions. The results of deleting gcnE or adaB on the expression of prnB and prnD are surprising. There is no effect on the noninduced (undetectable) and induced levels. However, a partial derepression of prnB and prnD is seen in both deletion strains. Previous work (10) has shown that only the repression of prnB is direct; the repression of prnD is, instead, the result of inducer exclusion. The latter easily accounts for the fact that the mild derepression of prnB results in an elevated derepression of prnD, particularly pronounced in the gcnEΔ strain. Interestingly, neither deletion significantly alters the induced levels of prnB or prnD transcripts, suggesting that the induction process could be independent of the histone acetylation status.
Chromatin organization in the prnB-prnD promoter is affected in SAGA/ADA component deletion strains.
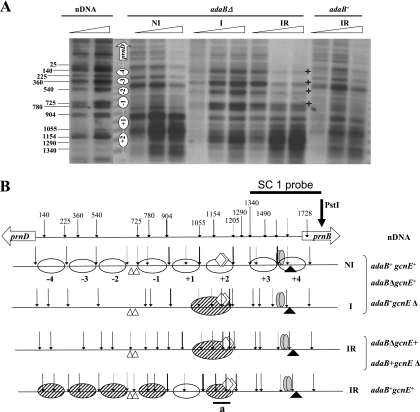
Figure 2A shows the results of an MNase digestion assay for adaB+ and adaBΔ under the three standard conditions investigated. The results of MNase digestions in the gcnEΔ strain (data not shown) are identical to those obtained for the adaB deletion. In both deletion strains, the only difference from the corresponding isogenic wild-type control in the nuclease digestion pattern is observed under inducing-repressing conditions. In an adaB+ gcnE+ strain, all nucleosomes are repositioned under inducing-repressing conditions (12), whereas in the adaBΔ and gcnEΔ strains, this repositioning is defective. In the deletion strains, the induced-repressed nucleosome pattern is identical to that seen under inducing conditions, which is also identical to the pattern obtained under inducing conditions in the wild-type strain. Hence, the derepressed expression of prnB and prnD observed in strains with deletions of adaB and gcnE is associated with and could depend on defective nucleosome repositioning. The SC1 probe shown here allows the detection of the region including that from nucleosome −4 to nucleosome +2. The probe detecting the region that includes nucleosomes +3 and +4 (probe SC2) did not reveal any differences between the deletion strains and the corresponding wild-type strains (data not shown). A graphical representation of nucleosome positioning in the prnB-prnD promoter region is shown in Fig. 2B. The nucleosome positioning in the adaBΔ strain under inducing-repressing conditions and a derepressed phenotype for prnB and prnD expression closely resemble the results obtained when the deacetylase inhibitor TSA is used in the wild type or when MNase patterns are analyzed in a creAd1 strain (10). We thus used ChIP to determine the acetylation status of histone H3 in the region of nucleosome +2, the only nucleosome that remains positioned under these conditions in the adaBΔ and gcnEΔ strains.
FIG. 2.
Nucleosome positioning in the prnD-prnB intergenic region in adaB+ and adaBΔ strains. (A) Indirect end-labeling MNase I analysis of the prnD-prnB promoter region using probe SC1. This probe reveals nucleosomes +2 to −4. MNase analysis of nucleosomes +3 and +4 is not shown as the patterns obtained for the mutant strains are identical to the pattern of the wild type. Growth conditions were identical to those described in the legend of Fig. 1: NI, noninducing; I, inducing; IR, inducing-repressing. Numbers beside the autoradiograms correspond to the positions of the main cuts relative to the prnD ATG. These were calculated from molecular size markers run in every gel. Asterisks indicate the positions of the relevant changes observed. Triangles indicate increasing concentrations of MNase I. nDNA, naked DNA. To the left of the adaBΔ NI lanes, a schematic representation of the nucleosome structure is shown. The wild-type patterns under all conditions have been previously published (12); we include in the same gel for comparison only the pattern obtained under inducing-repressing conditions, the only one where an adaB deletion pattern differs from that of the wild type. (B) Schematic representation of nucleosome positioning (based on the results shown in panel A and data from reference 12). All eight nucleosomes of the intergenic region are drawn. Arrows indicate MNase I digests. Their thicknesses indicate the relative intensities of the bands in the autoradiogram shown in panel A and in other autoradiograms (data not shown) covering nucleosomes +3 and +4. Dashed arrows indicate weakly cut sites. Under noninducing and inducing conditions, the nucleosome patterns are identical in the adaB+ and adaBΔ strains. White ovals represent fully positioned nucleosomes, while partially positioned nucleosomes are shown by diagonally hatched ovals. The interpretation and significance of this partial positioning are discussed in the text (see also references 12 and 25). Symbols: white lozenges, CreA-binding sites 3.1 and 3.2, which are mutated in the prnd20 and prnd22 strains and result in derepression (see text); gray ovals, AreA-binding sites 13 and 14, shown to be the physiologically relevant AreA binding sites (15); white triangles, high-affinity PrnA binding sites 2 and 3 (14); black triangle, TATA box (15). The fragment amplified in the ChIP analysis (fragment a) is shown as a solid line below nucleosome +2. nDNA, pattern obtained with naked DNA. To the right of the scheme we indicate the strain(s) of the corresponding nucleosome pattern.
The acetylation status of histone H3 in the region of nucleosome +2 depends on physiological conditions and on the function of GcnE and AdaB.
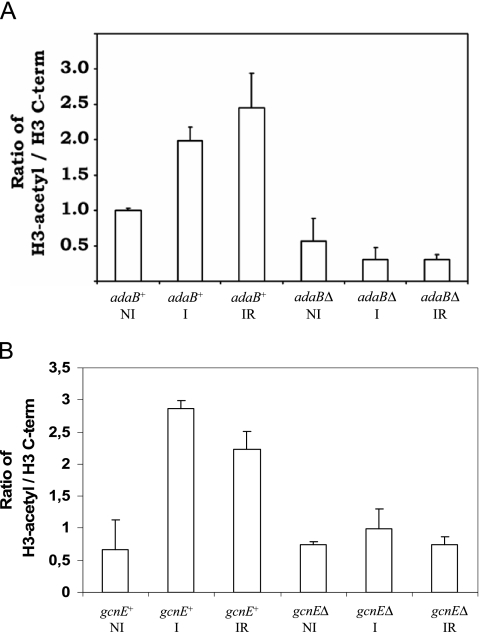
We chose to look at the region of nucleosome +2, as MNase digests indicate that a nucleosome is present in this region under all conditions, whether completely positioned (noninducing conditions) or partially positioned (inducing and inducing-repressing conditions) (12). The acetylation status of H3K9 and H3K14 is a critical mark for gene activation. To investigate the acetylation status of H3K9 and H3K14 in nucleosome +2, we employed ChIP assays using an antibody recognizing both acetyl-H3K9 and acetyl-H3K14, followed by the amplification of a fragment encompassing the region covered by nucleosome +2 (Fig. 2B, fragment a). The ratio between acetylated and total H3 (precipitated with an antibody directed against an H3 C-terminal epitope) is shown in Fig. 3. The acetylation patterns in the adaBΔ and gcnEΔ strains are very similar, but, in contrast to the MNase digests, they are not identical and are therefore presented here for both strains.
FIG. 3.
ChIP assay comparing acetylated H3K9 and -K14 in nucleosome +2 of the prnD-prnB intergenic region between adaB+ and adaBΔ (A) and between gcnE+ and gcnEΔ (B) strains (see Table 1 for genotype details). The ratio between total H3 (C-terminal [C-term] epitope) and H3-acetyl (recognizing H3 K9/K14) in the adaB+ wild-type strain grown under noninducing conditions (NI) was set to 1. Error bars indicate the standard deviation of four experiments (two biological and two technical repetitions per condition). Growth conditions: NI, noninducing; I, inducing; IR, simultaneously inducing-repressing.
In the adaB+ (wild type) strain, isogenic to the adaBΔ strain, the acetylation of H3K9 and H3K14 increases with induction, indicating a correlation between H3 acetylation, chromatin rearrangements (loss of positioning), and transcriptional activation. Upon repression, the acetylation status of H3K9 and H3K14 does not change significantly, which indicates that the deacetylation of these residues in H3 is not required to trigger gene repression and repositioning of nucleosome +2. In contrast to adaB+, in the adaBΔ strain the acetylation of H3K9 and H3K14 is strongly reduced under all conditions; and despite considerable induction of prnB and prnD transcription, an increase in the acetylation of H3 is not seen in these strains. A qualitatively very similar pattern is observed when the gcnE+ (wild type) control strain is compared to the isogenic gcnEΔ strain under the different growth conditions. As in the adaBΔ strain, the acetylation of H3K9 and H3K14 is strongly reduced in the gcnE deletion strain under all conditions.
DISCUSSION
Often, enhanced gene expression correlates with loss of positioning and increased acetylation of nucleosomes (33). In the prnD-prnB bidirectional promoter, we see increased acetylation of H3K9 and H3K14 under inducing conditions in the region corresponding to partially positioned nucleosome +2. However, this increased acetylation is not required for efficient transcription or chromatin remodeling. In the strains lacking essential components of the putative SAGA/ADA complexes, i.e., GcnE or AdaB, we detect only low levels of H3 lysine acetylation, but the induction and remodeling of chromatin are not affected. This is not without precedence, as in S. cerevisiae, SAGA/ADA-dependent and -independent activators are known. The activation of transcription requires SAGA or ADA complexes in the GAL1 promoter where GAL4 recruits the SAGA complex, and this in turn is necessary for the recruitment of the mediator complex (20). Our results indicate that, while a putative SAGA or ADA complex might be involved in determining the acetylation levels in this region, the PrnA-mediated activation and chromatin remodeling of the prnD-prnB promoter do not depend on AdaB and GcnE, putative members of A. nidulans SAGA/ADA complexes.
It should be noted that in our ChIP experiments, we have tested only histone H3 acetylation at lysines 9 and 14, residues known to be acetylated by SAGA and ADA complexes (9). Other covalent modifications not detected by the antibody used here, such as acetylations of H2A, H2B, and H4 at diverse lysines in their N termini, could also lead to the hyperacetylation of nucleosomes in the prnD-prnB promoter, thus providing the signal for effective chromatin remodeling. In filamentous fungi, it is completely unknown which mechanisms direct the cross talk of histone modification. In a study focusing on the different roles of H3 and H4 acetylation in the yeast S. cerevisiae, it was shown that H4 hyperacetylation in promoters of Adr1p-dependent genes does not compensate for low H3 acetylation in a gcn5 mutant strain (1).
The observed partial derepression of prnB and prnD expression in strains with deletions of adaB and gcnE was unexpected. However, in N. crassa, mutation of the GCN5 homologue ngf-1 results in loss of repression of the blue-light-responsive al-3 gene in the dark (18). An ngf-1RIP1 mutant was found to express al-3 constitutively, suggesting that the HAT is involved not only in blue-light induction but also in the repression of al-3 in the absence of light. Another example of HAT activities involved in both gene activation and repression is provided by the yeast GAL system. Glucose repression requires ADA2p and ADA3p/NGG1p, an ADA complex protein functionally associated with GCNp5 and ADA2p (6), and it has been suggested that ADA2p/ADA3p directly inhibits the activation domain of GAL4p. Similarly, in the yeast arginine metabolic system, GCN5 function and increased histone acetylation are associated with transcriptional repression. In this case, a Gcn5p-mediated increase of H3 acetylation correlates with binding of the arginine repressor complex ARGR/MCM1 (31).
However, our nucleosome-positioning and ChIP studies lead to a different picture. Although histone H3 acetylation increases during transcriptional activation, it is not required for transcriptional activation (compare results of inducing conditions in gcnE+/adaB+ strains with those for gcnEΔ or adaBΔ strain in Fig. 1 and 3) or loss of nucleosome positioning (Fig. 2). Additionally, transcriptional repression and repositioning of nucleosomes are not accompanied by deacetylation of H3K9 or H3K14 (Fig. 3, IR lanes). These results suggest that differences in histone acetylation are associated with, but not necessary for, transcriptional activation and nucleosome-positioning processes in the prnD-prnB intergenic region. It was therefore surprising to see that both hyperacetylation by TSA inhibition of deacetylases and hypoacetylation by deletion of adaB or gcnE lead to partial derepression of the proline catabolic genes and a simultaneous lack of nucleosome repositioning.
Transcriptional repression and nucleosome repositioning upon simultaneous glucose and ammonium repression require a functional CreA protein. This has been shown by the results of mutations in both the CreA protein itself (creAd1) and in its cognate binding sites (prnd20 or prnd22) in the prnD-prnB promoter (12). The simplest hypothesis integrating our experimental results would therefore propose that both hyperacetylation by TSA treatment and hypoacetylation by adaB or gcnE deletion impair CreA function. Several observations support this view. First, the nucleosomal patterns obtained in the prnD-prnB region by TSA treatment and in adaBΔ and gcnEΔ under inducing-repressing conditions are identical to those found in creAd1, prnd20, or prnd22 mutant strains (13). Second, derepression under TSA treatment or in adaBΔ and gcnEΔ strains is only partial, whereas derepression is complete in creA loss-of-function strains. This also implies that while nucleosome repositioning may be necessary for full repression, CreA can still partially repress on completely open chromatin, probably by directly interfering with the activating function of the pathway-specific activator PrnA.
Third, the function of CreA, but not the acetylation status of histones, defines whether nucleosomes are positioned or not. In summary, nucleosomal rearrangements, transcriptional activity, and histone acetylation are clearly distinct processes in the prnD-prnB region, and therefore a direct influence of the acetylation status on nucleosomal positioning and transcription is highly unlikely. Further studies will reveal at which level, transcriptional and/or posttranscriptional, CreA function is impaired by TSA treatment or the lack of HAT activities.
Supplementary Material
Acknowledgments
We are grateful for receiving strain MH 9233 from Michael Hynes (The University of Melbourne).
This work was supported by grant P17018 of the Austrian Science Fund FWF to J.S. Work at Orsay was supported by the CNRS, the Université Paris-Sud, and the Institut Universitaire de France. Y.R.-D. was supported by a predoctoral studentship of the Ministère de l'Education Supérieure et de la Recherche and the CONACYT (México). R.F.-M. was supported by a Marie Curie Fellowship (MCFI-2001-01084). I.G. was supported by postdoctoral fellowships from, successively, the Spanish Ministry of Education and the EU Marie Curie Programme.
Footnotes
Published ahead of print on 22 February 2008.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Agricola, E., L. Verdone, E. Di Mauro, and M. Caserta. 2006. H4 acetylation does not replace H3 acetylation in chromatin remodelling and transcription activation of Adr1-dependent genes. Mol. Microbiol. 621433-1446. [DOI] [PubMed] [Google Scholar]
- 2.Balasubramanian, R., M. G. Pray-Grant, W. Selleck, P. A. Grant, and S. Tan. 2002. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J. Biol. Chem. 2777989-7995. [DOI] [PubMed] [Google Scholar]
- 3.Berger, H., R. Pachlinger, I. Morozov, S. Goller, F. Narendja, M. Caddick, and J. Strauss. 2006. The GATA factor AreA regulates localization and in vivo binding site occupancy of the nitrate activator NirA. Mol. Microbiol. 59433-446. [DOI] [PubMed] [Google Scholar]
- 4.Bernreiter, A., A. Ramon, J. Fernandez-Martinez, H. Berger, L. Araujo-Bazan, E. A. Espeso, R. Pachlinger, A. Gallmetzer, I. Anderl, C. Scazzocchio, and J. Strauss. 2007. Nuclear export of the transcription factor NirA is a regulatory checkpoint for nitrate induction in Aspergillus nidulans. Mol. Cell. Biol. 27791-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhaumik, S. R., and M. R. Green. 2001. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 151935-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandl, C. J., J. A. Martens, A. Margaliot, D. Stenning, A. M. Furlanetto, A. Saleh, K. S. Hamilton, and J. Genereaux. 1996. Structure/functional properties of the yeast dual regulator protein NGG1 that are required for glucose repression. J. Biol. Chem. 2719298-9306. [DOI] [PubMed] [Google Scholar]
- 7.Brownell, J. E., J. Zhou, T. Ranalli, R. Kobayashi, D. G. Edmondson, S. Y. Roth, and C. D. Allis. 1996. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84843-851. [DOI] [PubMed] [Google Scholar]
- 8.Candau, R., J. X. Zhou, C. D. Allis, and S. L. Berger. 1997. Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo. EMBO J. 16555-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carrozza, M. J., R. T. Utley, J. L. Workman, and J. Cote. 2003. The diverse functions of histone acetyltransferase complexes. Trends Genet. 19321-329. [DOI] [PubMed] [Google Scholar]
- 10.Cubero, B., D. Gomez, and C. Scazzocchio. 2000. Metabolite repression and inducer exclusion in the proline utilization gene cluster of Aspergillus nidulans. J. Bacteriol. 182233-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cubero, B., and C. Scazzocchio. 1994. Two different, adjacent and divergent zinc finger binding sites are necessary for CREA-mediated carbon catabolite repression in the proline gene cluster of Aspergillus nidulans. EMBO J. 13407-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia, I., R. Gonzalez, D. Gomez, and C. Scazzocchio. 2004. Chromatin rearrangements in the prnD-prnB bidirectional promoter: dependence on transcription factors. Eukaryot. Cell 3144-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomez, D., B. Cubero, G. Cecchetto, and C. Scazzocchio. 2002. PrnA, a Zn2Cys6 activator with a unique DNA recognition mode, requires inducer for in vivo binding. Mol. Microbiol. 44585-597. [DOI] [PubMed] [Google Scholar]
- 14.Gomez, D., I. Garcia, C. Scazzocchio, and B. Cubero. 2003. Multiple GATA sites: protein binding and physiological relevance for the regulation of the proline transporter gene of Aspergillus nidulans. Mol. Microbiol. 50277-289. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez, R., V. Gavrias, D. Gomez, C. Scazzocchio, and B. Cubero. 1997. The integration of nitrogen and carbon catabolite repression in Aspergillus nidulans requires the GATA factor AreA and an additional positive-acting element, ADA. EMBO J. 162937-2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez, R., and C. Scazzocchio. 1997. A rapid method for chromatin structure analysis in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 253955-3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grant, P. A., L. Duggan, J. Cote, S. M. Roberts, J. E. Brownell, R. Candau, R. Ohba, T. Owen-Hughes, C. D. Allis, F. Winston, S. L. Berger, and J. L. Workman. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 111640-1650. [DOI] [PubMed] [Google Scholar]
- 18.Grimaldi, B., P. Coiro, P. Filetici, E. Berge, J. R. Dobosy, M. Freitag, E. U. Selker, and P. Ballario. 2006. The Neurospora crassa White Collar-1 dependent blue light response requires acetylation of histone H3 lysine 14 by NGF-1. Mol. Biol. Cell 174576-4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koehler, R. N., N. Rachfall, and R. J. Rolfes. 2007. Activation of the ADE genes requires the chromatin remodeling complexes SAGA and SWI/SNF. Eukaryot. Cell 61474-1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koh, S. S., A. Z. Ansari, M. Ptashne, and R. A. Young. 1998. An activator target in the RNA polymerase II holoenzyme. Mol. Cell 1895-904. [DOI] [PubMed] [Google Scholar]
- 21.Kuo, M. H., J. E. Brownell, R. E. Sobel, T. A. Ranalli, R. G. Cook, D. G. Edmondson, S. Y. Roth, and C. D. Allis. 1996. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature 383269-272. [DOI] [PubMed] [Google Scholar]
- 22.Larschan, E., and F. Winston. 2001. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 151946-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, T. I., H. C. Causton, F. C. Holstege, W. C. Shen, N. Hannett, E. G. Jennings, F. Winston, M. R. Green, and R. A. Young. 2000. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature 405701-704. [DOI] [PubMed] [Google Scholar]
- 24.Lee, T. I., and R. A. Young. 2000. Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet. 3477-137. [DOI] [PubMed] [Google Scholar]
- 25.Mathieu, M., I. Nikolaev, C. Scazzocchio, and B. Felenbok. 2005. Patterns of nucleosomal organization in the alc regulon of Aspergillus nidulans: roles of the AlcR transcriptional activator and the CreA global repressor. Mol. Microbiol. 56535-548. [DOI] [PubMed] [Google Scholar]
- 26.Monahan, B. J., J. A. Fraser, M. J. Hynes, and M. A. Davis. 2002. Isolation and characterization of two ammonium permease genes, meaA and mepA, from Aspergillus nidulans. Eukaryot. Cell 185-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muro-Pastor, M. I., R. Gonzalez, J. Strauss, F. Narendja, and C. Scazzocchio. 1999. The GATA factor AreA is essential for chromatin remodelling in a eukaryotic bidirectional promoter. EMBO J. 181584-1597. (Erratum, 18:2670.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muro-Pastor, M. I., J. Strauss, A. Ramon, and C. Scazzocchio. 2004. A paradoxical mutant GATA factor. Eukaryot. Cell 3393-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Narendja, F., S. P. Goller, M. Wolschek, and J. Strauss. 2002. Nitrate and the GATA factor AreA are necessary for in vivo binding of NirA, the pathway-specific transcriptional activator of Aspergillus nidulans. Mol. Microbiol. 44573-583. [DOI] [PubMed] [Google Scholar]
- 30.Pontecorvo, G., J. A. Roper, L. M. Hemmons, K. D. MacDonald, and A. W. J. Bufton. 1953. The genetics of Aspergillus nidulans. Adv. Genet. 5141-238. [DOI] [PubMed] [Google Scholar]
- 31.Ricci, A. R., J. Genereaux, and C. J. Brandl. 2002. Components of the SAGA histone acetyltransferase complex are required for repressed transcription of ARG1 in rich medium. Mol. Cell. Biol. 224033-4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riego, L., A. Avendano, A. DeLuna, E. Rodriguez, and A. Gonzalez. 2002. GDH1 expression is regulated by GLN3, GCN4, and HAP4 under respiratory growth. Biochem. Biophys. Res. Commun. 29379-85. [DOI] [PubMed] [Google Scholar]
- 33.Roth, S. Y., J. M. Denu, and C. D. Allis. 2001. Histone acetyltransferases. Annu. Rev. Biochem. 7081-120. [DOI] [PubMed] [Google Scholar]
- 34.Sophianopoulou, V., and C. Scazzocchio. 1989. The proline transport protein of Aspergillus nidulans is very similar to amino acid transporters of Saccharomyces cerevisiae. Mol. Microbiol. 3705-714. [DOI] [PubMed] [Google Scholar]
- 35.Wang, L., C. Mizzen, C. Ying, R. Candau, N. Barlev, J. Brownell, C. D. Allis, and S. L. Berger. 1997. Histone acetyltransferase activity is conserved between yeast and human GCN5 and is required for complementation of growth and transcriptional activation. Mol. Cell. Biol. 17519-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.