Figure 6. Characterization of diap1 enhancers.
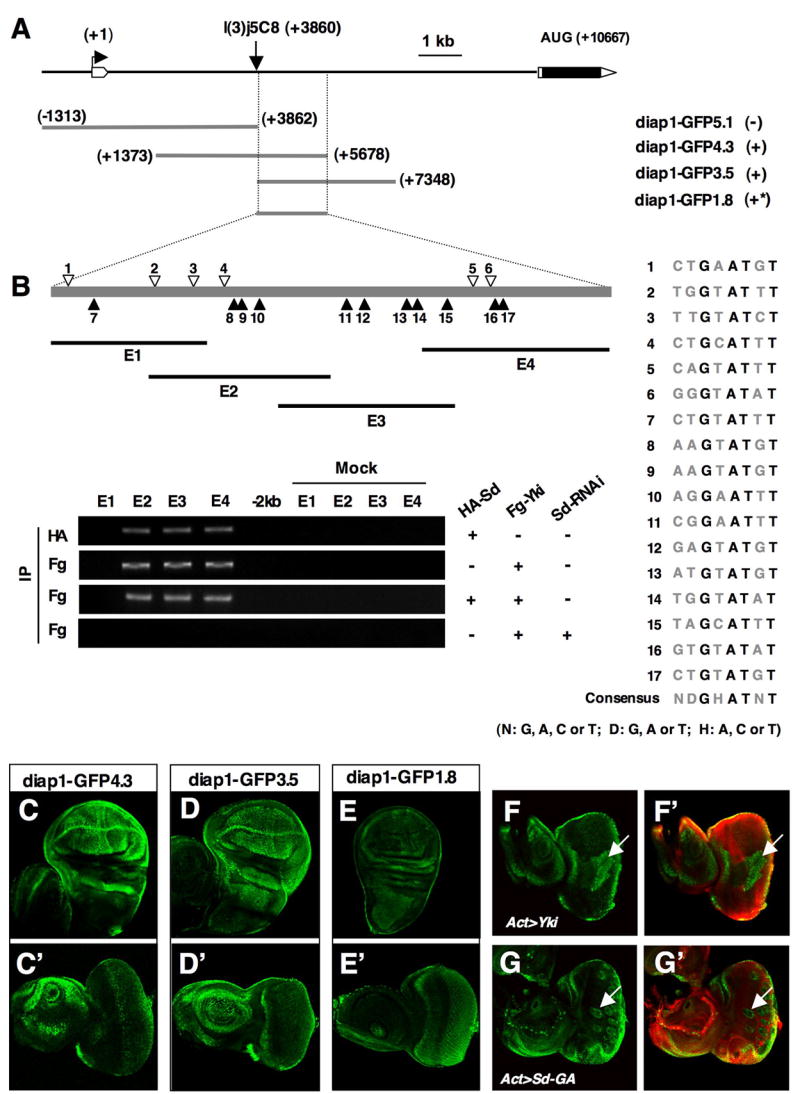
(A) Deletion analysis of diap1 enhancer elements. The diap1 genomic region containing the first intron and two neighboring exons is shown at the top. A diap1-lacZ enhancer trap line, l(3)j5C8, is inserted at +3860 bp. diap1-GFP transgenes containing indicated genomic fragments are listed. (−): no discernible expression; (*): weaker expression. (B) ChIP analysis of the 1.8 kb diap1 enhancer. Individual Sd binding consensus sites are indicated by open or filled arrowheads (filled arrowheads indicate the binding sites with reversed orientation). The sequences of individual Sd binding consensus sites are shown to the right. E1 to E4 demarcate the regions amplified by PCR in the ChIP experiments. (C–E′) GFP expression in wing (C, D, E) or eye (C′, D′, E′) discs driven by diap1-GFP4.3 (C, C′), diap1-GFP3.5 (D, D′), or diap1-GFP1.8 (E, E′). (F–G′) Eye discs that contain diap1-GFP4.3 and flip-out clones expressing Yki (F, F′) or Sd-GA (G, G′) with act>CD2>Gal4 were immunostained to the expression of GFP (green) and CD2 (red). Cells expressing Yki or Sd-GA were marked by the lack of CD2 expression.
