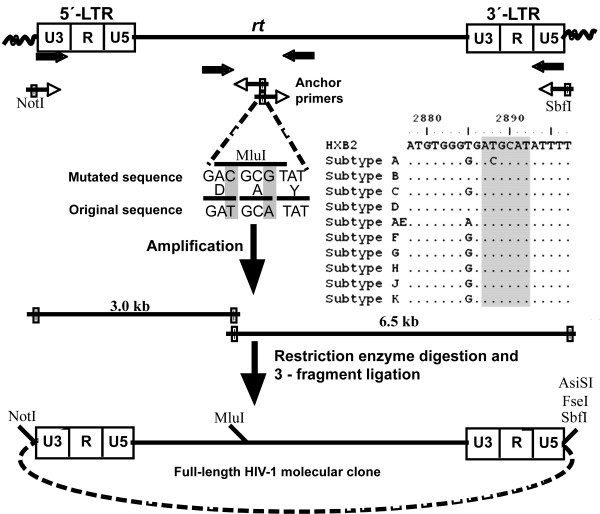
Figure 1.
Schematic representation of the anchor primer-based cloning strategy for HIV-1: An integrated provirus is depicted at the top of the cartoon with wavy lines representing the cellular genomic DNA. The figure is not drawn to the scale. The U3, R and U5 regions of both the LTRs are delineated. Arrows represent primer location and the orientation. The open arrows are used for the first-round and the filled arrows for the second round amplifications. Note that only the second-round primers contain restriction sites for cloning (filled boxes). The anchor primers, in both the orientations, are complementary to a highly conserved sequence within rt. The 6 nucleotide sequence into which the MluI site is engineered (the inset) and the flanking sequences where the anchor primers anneal (not completely shown) are highly conserved among diverse viral subtypes of HIV-1. Dots indicate sequence homology. The amino acid sequence is shown in the single letter code. The coordinates are as in HXB2 molecular clone (accession number K03455). The sequence into which MluI site was engineered is highlighted by shading. The complete viral genome is amplified in two unequal fragments and cloned directionally into a vector in a single ligation reaction to generate full-length molecular clones. Additional restriction sites, all 8 bp recognition sequences that can be used for the viral cloning are also shown.