Abstract
Impedance measurements of cell-based sensors are a primary characterization route for detection and analysis of cellular responses to chemical and biological agents in real time. The detection sensitivity and limitation depend on sensor impedance characteristics and thus on cell patterning techniques. This study introduces a cell patterning approach to bind cells on microarrays of gold electrodes and demonstrates that single-cell patterning can substantially improve impedance characteristics of cell-based sensors. Mouse fibroblast cells (NIH3T3) are immobilized on electrodes through a lysine-arginine-glycine-aspartic acid (KRGD) peptide-mediated natural cell adhesion process. Electrodes are made of three sizes and immobilized with either covalently-bound or physically-adsorbed KRGD (c-electrodes or p-electrodes). Cells attached to c-electrodes increase the measurable electrical signal strength by 48.4%, 24.2%, and 19.0% for three electrode sizes, respectively, as compared to cells attached to p-electrodes, demonstrating that both the electrode size and surface chemistry play a key role in cell adhesion and spreading and thus the impedance characteristics of cell-based sensors. Single cells patterned on c-electrodes with dimensions comparable to cell size exhibit well-spread cell morphology and substantially outperform cells patterned on electrodes of other configurations.
Keywords: cell-based sensor, impedance, peptide, cell patterning, cell adhesion, single cell
1. Introduction
Cell-based biosensors, also known as cytosensors, are hybrid systems that utilize living biological cells as sensing elements to monitor physiological changes induced by internal aberration or external stimuli, offering new opportunities for biomedical applications such as drug evaluation, biothreat detection, and environmental pollutant monitoring (Stett et al., 2003; Ziegler, 2000). They have the unique advantage over conventional chemical-, antibody-, or nucleic acid-based assays in that they provide insight into the physiological effect of an analyte and are capable of detecting both known and unknown biomolecules (McFadden, 2002; Stenger et al., 2001). Cytosensors are commonly addressed electronically through impedance characterization (impedance cytosensors). Impedance cytosensors offer instantaneous and quantitative means to study cellular events, such as changes of ionic channels in cell membrane, the variations of cell membrane integrity, and cell spreading, motility, and growth (Arndt et al., 2004; Kovacs, 2003) and detect analytes by transducing cellular responses into a measurable electrical signal (Asphahani and Zhang, 2007). Impedance cytosensors have been investigated to detect both toxic and noxious agents (Ceriotti et al., 2007; Gilchrist et al., 2005; Hartmann et al., 2007; Xiao and Luong, 2003) and monitor apoptosis-induced changes in cell shape (Arndt et al., 2004; Yin et al., 2007). However, in traditional impedance cytosensors, a large cell population is usually patterned over individual electrodes due to the limited ability to control cell adhesion processes, and thus the number of cells and the morphology of individual cells on one electrode are unpredictable. While averages of cell properties, such as proliferation, motility, and cell-cell separation, can be monitored over the cell population, it is impossible to examine individual cells and precisely monitor changes in cell properties. Importantly, with multiple cells on one electrode, the measurements of impedance changes as the cells respond to a stimulus become increasingly difficult. For example, the individual cells on one electrode may react differently, and the noise due to cell-cell interactions may suppress the acquired signal. Additionally, adhesion of multiple cells on an electrode often lead to greater signal loss because of the current leakage along the pathways between cells as a result of the direct exposure of the electrode to electrolyte (Huang et al., 2004). Therefore, to circumvent these limitations, single-cell based sensors may prove to be an appealing approach.
In this study, we use a ligand-mediated process to pattern single or multiple living cells on microelectrodes (Veiseh et al., 2007) and demonstrate the influence of the patterning process and electrode geometry on the impedance characteristics of the cell-based sensors. Electrodes of three different sizes were made and surface-modified with adhesion peptides (KRGD) through either physical adsorption or covalent bonding to mediate subsequent cell adhesion and spreading. The morphology of cells adhered on electrodes of different sizes and surface chemistries was examined, and the corresponding impedance characteristics of resulting cell-based sensors were studied.
2. Experimental
2.1 Materials
The following materials and chemicals were used as received: Nano-Strip™ 2X (Cyantek, Fremont, CA); 2-[methoxy(polyethyleneoxy)propyl]trimethoxysilane (Mw = 460–590 Da) (Gelest, Morrisville, PA); heat-inactivated fetal bovine serum (FBS), penicillin-streptomycin-neomycin (PSN) antibiotic 100X mixture, and Prolong® Gold antifade reagent with DAPI (Invitrogen, Carlsbad, CA); Dulbecco’s Modified Eagle Medium (DMEM), and 1X phosphate buffered saline solution (PBS) (Gibco, Carlsbad, CA); Alexa Fluor 594-conjugated wheat germ agglutinin (WGA; W11262) (Molecular Probes, Eugene, OR); lysine-arginine-glycine-aspartic acid (KRGD) oligopeptide (SynBioSci, Livermore, CA); 11-mercaptoundecanoic acid 95% (11-MUA), 3-mercaptopropionic acid 99% (3-MPA), N-hydroxysuccinimide 97% (NHS), 1-ethyl-3-(3-(dimethylamino)-propyl) carbodiimide (EDAC), trypsin-EDTA (ethylenediamine tetra-acetic acid), Sigmacote™, and glutaraldehyde and paraformaldehyde (Sigma-Aldrich, Milwaukee, WI). All the solvents including toluene and triethylamine were HPLC grade and were purchased from Aldrich (Milwaukee, WI). Absolute ethanol was always deoxygenated by dry N2 before use. NIH3T3 cells were obtained from the American Type Culture Collection (Manassas, VA).
2.2 Surface modification and cell culture
Microarrays of gold electrodes were patterned on silicon oxide substrates by conventional photolithography (see supplementary materials). Substrates were then modified following a previously established procedure with minor modifications (Veiseh et al., 2004; Veiseh et al., 2002). The gold/silicon oxide substrates were washed with acetone, ethanol, and DI water, respectively, to remove the protective photoresist layer, placed in NanoStrip™ 2X at room temperature for 30 min, rinsed thoroughly with DI water, and dried under nitrogen. The gold regions of the piranha-treated substrates were reacted with a 20 mM mixture of alkanethiols 11-mercaptoundecanoic acid (MUA) and 3-mercaptopropionic acid (MPA) (1:10 v/v) for 16 hours to form a self-assembled monolayer (SAM). The silicon background was passivated with methoxy-polyethyleneglycol-silane (methoxy-PEG-silane) solution prepared in nitrogen-filled reaction flasks by adding 3 mM PEG-silane in anhydrous toluene containing 1% triethylamine. The reaction proceeded under nitrogen at 60°C for 18 hrs. Loosely bound moieties were removed from the surfaces by sonicating in toluene and ethanol for 5 min each, followed by rinsing with DI water and drying under nitrogen. For surfaces to be covalently linked with KRGD, substrates were immersed in an aqueous solution of 150 mM EDAC and 30 mM N-hydroxysuccinimide (NHS) for 30 min to attach the NHS ester intermediate to activate carboxylate groups of the alkanethiol SAM to chemically bond primary amino groups of KRGD, where the lysine residues displace the NHS group during the reaction. For substrates to be linked with physically adsorbed KRGD, NHS/EDAC was not used. The substrates were then sterilized with absolute ethanol for 15 min and exposed to KRGD peptide at a concentration of 50 μg/mL in PBS of 8.2 pH at room temperature for 1 hr. To remove loosely bound moieties after each step of the surface modification, the substrate was rinsed with its original solvent and DI water, respectively.
Mouse fibroblast (NIH3T3) cell line was cultured in 75 cm2 flasks at 37°C in a humidified atmosphere with 5% CO2. The cell medium contains 10% FBS and 1% PSN antibiotic in DMEM supplemented with 4 mM L-glutamine, 1.5 g/L sodium bicarbonate, and 4.5 g/L glucose. The medium was changed every third day. For cell adhesion, 0.5 mL of NIH3T3 cells at a concentration of 2 × 105 cells/mL was plated onto the peptide-patterned substrates. The cells were allowed to adhere to the substrates for 24 hours under the standard culture conditions. After electrical measurements were completed, the cells were fixed with Karnovsky’s fixative for 30 minutes at room temperature for optical imaging using a differential interference contrast (DIC) reflectance microscope (Nikon E800 Upright Microscope, New York, NY).
2.3 Fluorescence imaging of cell membranes and nuclei
Cell-patterned substrates were fixed with 4% paraformaldehyde solution. Following fixation, cells were stained with Alexa Fluor 594-conjugated WGA (membranes) and 4′,6-diamidino-2-phenyindole (DAPI) (nuclei) according to the manufacturers’ instructions. Confocal images were acquired using a DeltaVision SA3.1 wide-field deconvolution microscope (Applied Precision, Inc., Issaquah, WA) equipped with DAPI and TRITC.
2.4 Instrumentation and electrical characterization of cells on electrode arrays
An Agilent 33220A waveform generator was used as the voltage source (Palo Alto, CA). Electronic measurements were performed using a Stanford Research Systems SR-510 lock-in amplifier (Sunnyvale, CA) and 1 μm tip tungsten probes obtained from Signatone (Gilroy, CA).
The integrated microelectrode array (IMA) patterned with NIH3T3 cells were retrieved from cell culture and inspected under an optical reflectance microscope to ensure the cell attachment to the detecting electrodes. Electrolyte solution was removed from the measurement pads at the edges of the IMA chips. IMA chips were placed in an insulative holder that resided within a probe station to record electrical measurements. The input signal of a 100 mV (peak-to-peak) sine wave at a frequency of 4 kHz was passed through a 1 MΩ resistor to limit the current amplitude to 100 nA. The alternating current potential was applied between the detecting and counter electrodes by connecting tungsten probes to the measurement pads of the IMA chip. Electrical characterization involved measuring the voltage amplitude between the detecting electrode and counter electrode (ΔV) and the signal phase shift (Δϕ) using a lock-in amplifier.
3. Results and discussion
3.1 Cell adhesion on microelectrode arrays
Because cell adhesion triggers signals that regulate normal cell functions such as growth, differentiation, and motility (Hynes, 1992), it is preferable to pattern cells onto microelectrodes through natural cell adhesion rather than mechanical positioning, which helps retain cellular viability (Veiseh and Zhang, 2006). This was accomplished in this study by immobilizing KRGD peptides on microarrays of electrodes to mediate the subsequent NIH3T3 cell adhesion and spreading. NIH3T3 cells were patterned on square electrodes of three different sizes (25 μm, 30 μm, and 110 μm in side length) that were chemically modified with either covalently-bound (c-electrodes) or physically-adsorbed (p-electrodes) KRGD. Here, electrodes with physically-adsorbed KRGD served as control in this comparison study, as such a configuration has been widely used to promote cell attachment and spreading (Luong, 2003). To inhibit protein adsorption and cell adhesion on the insulative background, a layer of methoxy-PEG-silane was deposited onto the silicon oxide regions. Optical images in Figure 1A shows NIH3T3 cells adhered on c-electrodes of three different sizes. Fluorescent images in Figure 1B illustrate the difference in cell adhesion and spreading between NIH3T3 cells patterned on c-electrodes and p-electrodes.
Figure 1.
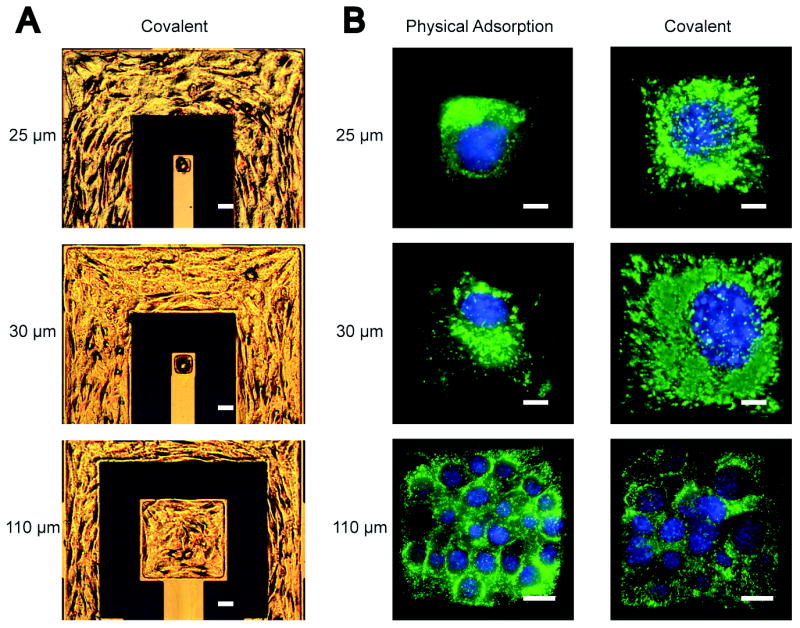
(A) Epi-DIC images of NIH3T3 single cells patterned on 25 μm and 30 μm detecting electrodes and multiple cells patterned on a 110 μm detecting electrode modified with covalently-bound KRGD. (B) Fluorescent images of nuclei- (blue) and membrane- (green) stained NIH3T3 cells on electrodes of three different sizes and modified with physically-absorbed (left, p-electrode) or covalently-bound (right, c-electrode) KRGD. The scale bar is 25 μm in (A), and 5 μm for 25-μm and 30-μm electrodes and 20 μm for 110-μm electrodes in (B).
Cellular membrane fluorescent images show that cells are poorly spread on the surfaces of p-electrodes (25 μm and 30 μm). Cell spreading is governed by the availability of adhesion sites on the surface (Veiseh et al., 2007). Due to weak binding between KRGD molecules and p-electrodes, physically-adsorbed KRGD molecules are vulnerable to detachment from the electrode upon exposure to cell culture medium and during the cell spreading process, an event that reduces the effective binding sites for cell spreading across the electrode (Veiseh et al., 2007). The cell on the c-electrode exhibits well-spread conformation and covers the entire surface area of the electrode as a result of strong covalent bonding of KRGD molecules to the electrode surface which provides a large number of robust adhesion sites for cell spreading. On the larger electrodes (110 μm), multi-cell adhesion was observed. However, much fewer cells were seen on the c-electrode than on the p-electrode due to the higher degree of cell spreading on the former.
3.2 Electric voltage measurements
A primary objective in development of cell-based sensors is to reduce the electrical signal loss resulted from current leakage along undesired shunt pathways (e.g. direct electrode-to-electrolyte exposure), thereby increasing the signal-to-noise ratio (SNR). Figure 2 shows the measured voltage magnitudes of electrode arrays before and after cells were patterned on the electrodes. Prior to cell patterning, baseline (i.e. cell-free) voltage and phase measurements were made in cell culture medium solution only. Voltage and phase data were then recorded for NIH3T3 cells adhered on the electrodes. Only voltage magnitude data is presented here since no appreciable phase shifts were observed at the source signal frequency of 4 kHz; the resistivity dominates the impedance measurements for the electrode/cell configurations presented in this study. The signal magnitude for cells adhered on c-electrodes of 25 μm, 30 μm, and 110 μm in side length shows an increase of 48.4%, 24.2%, and 19.0%, respectively, compared to cells adhered on p-electrodes of the same sizes. The result also indicates that the signal enhancement is more significant for electrodes of smaller sizes.
Figure 2.
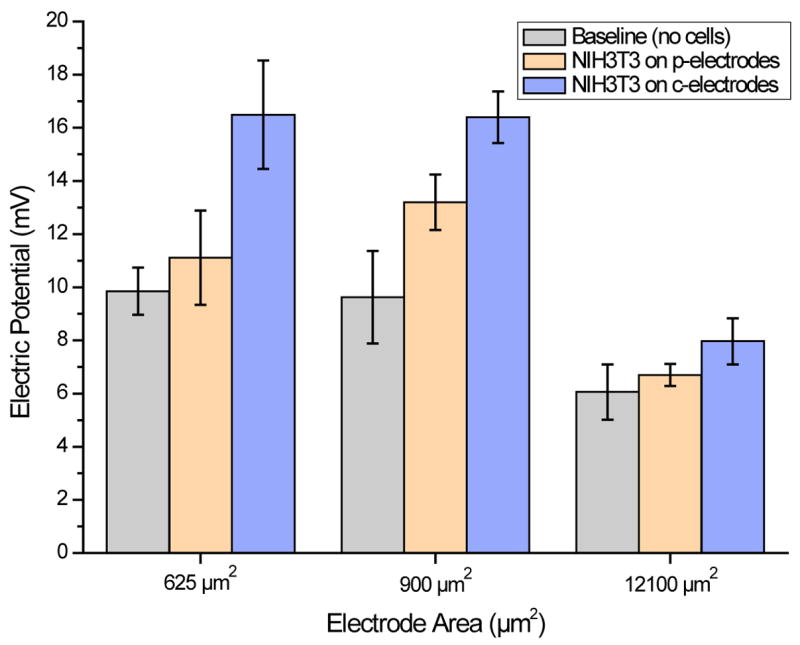
Voltage magnitudes of cell-free and NIH3T3 cell-covered electrodes with surface areas of 625 μm2 (25 μm × 25 μm), 900 μm2 (30 μm × 30 μm), and 12100 μm2 (110 μm × 110 μm). The electrodes were pre-coated with either covalently-bound (c-electrodes) or physically-absorbed KRGD peptides.
3.3 Circuit model
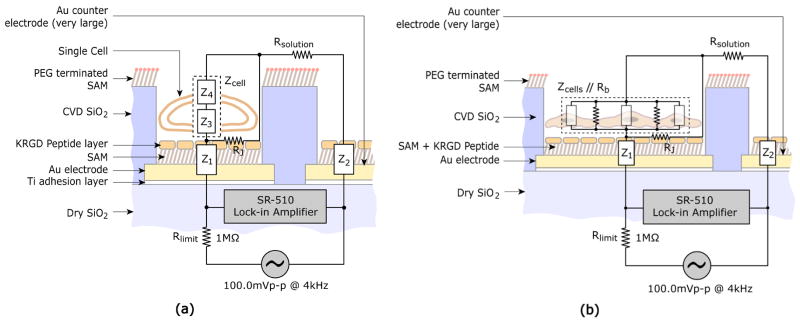
To determine the cellular impedance values from the measured voltage data, a circuit model of the cell-electrode heterostructure was constructed, as shown in Figure 3. A point-contact model is used to represent the cell-electrode interface or junction (Fromherz, 1999). Z1 and Z2 represent electrode-peptide-electrolyte interfacial impedances at the detecting and counter electrodes respectively, and cellular impedance, Zcell, is designated as the combination of the top membrane impedance, Z3, and the bottom membrane impedance, Z4 (Zcell = Z3 + Z4). The cytoplasmic impedance is neglected compared to the membrane impedance of a normal cell (Mossop et al., 2004; Omori et al., 2006). Because a large portion of the outer and periphery regions of the electrode are often separated from the cell membrane by a cleft filled with electrolyte of culture medium, a finite seal resistance or cell-to-substrate resistance exists and is represented by RJ in the model. Rsolution represents the resistance of the electrolyte solution, whose value is dependent on the ion concentration in the solution. In the practical designs of cell-based sensors, counter electrodes often are designed to have much larger surface areas than detecting electrodes (e.g. Fig. 1A), resulting in a Z2 much smaller than the cellular impedance Zcell and the surface interfacial impedance of the detecting electrodes Z1, thus enhancing the detection sensitivity (Giaever and Keese, 1991; Lo et al., 1995). The impedance of the culture medium (Rsolution) is also substantially lower than Z1 and Z2 due to the electrolyte nature of the culture medium solution. For multi-cell adhesion on larger detecting electrodes (e.g. Fig 1B), current that passes through the gaps between neighboring cells is taken into account by adding the total junctional resistance between neighboring cells (Rb), which is in parallel to total impedance of cells (Zcells).
Figure 3.
Circuit model and electric impedance characterization of cell-based sensors: (a) single-cell impedance measurements and (b) multi-cell impedance measurements. Z1: electrode-SAM-peptide impedance, Z2: counter-electrode-SAM-peptide impedance, Z3: cell membrane impedance (bottom), Z4: cell membrane impedance (top), Zcell: total single-cell impedance (Z3 + Z4), Rb: total resistance between neighboring cells, Zcells: impedance of cells, Rb//Zcells: impedance of monolayer of cells, RJ: cleft/seal resistance, Rsolution: solution resistance, and Rlimit: limiting resistance.
The cleft between the cell membrane and electrode surface is formed due to the finite binding force and the presence of the molecules that protrude from the cell membrane (e.g. integrins, glycocalix), keeping the lipid core of the membrane at a certain distance from the surface. This electrolyte-filled cleft, combined with the surface area uncovered by the cell, constitutes a shunt path with a “seal resistance” RJ bypassing the cell on the electrode. Clearly, if this seal resistance is too small compared to Zcell (i.e., RJ ≪ Zcell) due to a large cleft (Kataoka et al., 2002) or uncovered surface area, RJ dominates the impedance of the parallel circuit and the variation of the impedance due to changes in cell properties may not be readily detected. Additionally, the resulting measured voltage will be small due to the combined low resistance. Thus cell adhesion and spreading directly affect the impedance properties of cell-based sensors, and a tight cell binding to the electrode and a high surface coverage of the electrode by the cell may increase RJ and thus the sensitivity and detectability.
Using the circuit model, single cell (and multiple cells) related impedance values are determined from the experimentally measured voltage values (e.g. Fig. 2). The overall impedance of the circuit for a single-cell patterned electrode can be expressed as:
| (1) |
For multiple cells attached to a single electrode, the overall impedance can be formulated as follows:
| (2) |
The counter electrode impedance Z2 are negligible in equations (1) and (2) compared to the rest of the terms, because of its large surface area (Greve et al., 2003; Huang et al., 2004).
The probe current of the sensing circuit was clamped to 100 nA by the 1 MΩ limiting resistor (Fig. 3). The baseline impedance or cell-free electrode impedance can be calculated from the following formula:
| (3) |
where Vbaseline is the measured voltage across the cell-free detecting electrode and counter electrode and RLimit is the current-limiting resistor. The computation of the electrode/cell impedance, Zcell(s)–electrode, and the related cleft/seal resistance, RJ, is implemented by assuming a uniform sheet resistance across the bottom of the entire cell spreading over the detecting electrode and incorporating the shunt-connection between the membrane impedances. Zcell(s)–electrode values are evaluated using the respective measured voltages across the detecting electrode and counter electrode (Vmeasured) using the following formula:
| (4) |
After Zcell(s)-electrode and baseline impedance are obtained, RJ values are calculated using equations (1) and (2). Combined cell-and-cleft impedance values are calculated by subtracting Zcell-free from Zcell(s)-electrode. The difference between the cell-free electrode impedance and cell-covered electrode impedance represents the combined cell impedance and cleft/seal resistance.
3.4 Impedance of cell-electrode hetero-structure
In this study, by using electrodes with dimensions comparable to cell size and the natural cell adhesion mediated by adhesion ligands covalently bound on electrodes, single cell patterns with tight binding and full cell spreading over the entire electrode surface were achieved (Fig. 1), which effectively reduced electrode surface area exposed to electrolytic buffer and increased signal intensity. Using the circuit model presented in Figure 3 and the equations above, the values of cell-free electrode impedances, cell-covered electrode impedances, the values of percent changes in impedance (i.e., (Zcell(s)-electrode – Zcell-free)/Zcell-free) and cleft/seal resistance per unit electrode area (RJ/Aelectrode) were derived from the measured data (i.e. Vbaseline and Vmeasured) as shown in Table 1. Significantly higher seal resistance, RJ, is achieved for cells adhered on c-electrodes than cells on p-electrodes, as expected, and RJ increases as the electrode size decreases. Particularly, for the electrode with a side length of 25 μm, the difference is more than 10 fold. The significant increase in seal resistance on c-electrodes can be attributed to (1) covalently-bound adhesion ligands providing more robust binding sites for cell adhesion (Veiseh et al., 2007) and thus decreasing the distance between the cell membrane and the electrode, and (2) the well-spread cells (Fig. 1) reducing the uncovered electrode surface area exposed to the electrolyte and thus the current leakage. It should be noted that the difference between the cell-covered electrode impedance and cell-free electrode impedance, i.e., Zcell(s)-electrode – Zcell-free, is the impedance actually attributable to the adhered cell. This impedance difference should be large enough compared to the baseline impedance (Zcell-free) to identify the presence or absence of the cell on the electrode and to detect small changes on cell properties. For example, the single-cell electrodes exhibit substantial impedance changes upon cell adhesion (Supplementary Fig. 2A), while the multi-cell p-electrode exhibits an impedance change of only 3.8% increase above the baseline, which may be too small compared to the baseline impedance to obtain reliable measurements. Additionally, the impedance changes due to cell adhesion are substantially larger for the c-electrodes than the corresponding p-electrodes of the same sizes. Noticeably, the impedance change of the c-electrode due to cell adhesion is approximately 5.6 times larger than the impedance change of the p-electrode at the electrode size of 25 μm. Similarly, the electrode size influences the seal resistance per unit area, and the seal resistance of the c-electrode is approximately 11.8 times larger than that of the ps-electrode at the electrode size of 25 μm (Supplementary Fig. 2B).
Table 1.
Estimated impedance values of cell-electrode hetero-structure for different electrode dimensions and surface chemistries.
| Impedances Vs. Electrode Dimensions | 25 × 25 μm2 | 30 × 30 μm2 | 110 × 110 μm2 | |||
|---|---|---|---|---|---|---|
| Physical Adsorption | Covalent Bonding | Physical Adsorption | Covalent Bonding | Physical Adsorption | Covalent Bonding | |
| Cell-free Electrode Impedance (Z1 + Z2 + Rsolution or Zcell-free) | 109.3 ± 10.9 kΩ | 106.7 ± 21.3 kΩ | 73.6 ± 13.5 kΩ | |||
| Cell-covered Electrode Impedance (Zcell(s)-electrode) | 125.0 ± 22.4 kΩ | 197.5 ± 29.2 kΩ | 152.1 ± 13.8 kΩ | 196.0 ± 13.9 kΩ | 76.4 ± 5.1 kΩ | 96.5 ± 11.5 kΩ |
| Percent Change in Impedance (Zcell(s)-electrode – Zcell-free)/Zcell-free) | 14.4 ± 10.2 % | 80.7 ± 12.7 % | 42.5 ± 10.4 % | 83.7 ± 10.4 % | 3.8 ± 5.9 % | 31.1 ± 7.2 % |
| Averaged Cleft/Seal Resistance Per unit Electrode Area (RJ/Aelectrode) | 2.7 ± 0.8 GΩ/cm2 | 32.0 ± 6.0 GΩ/cm2 | 4.8 ± 0.8 GΩ/cm2 | 30.2 ± 4.2 GΩ/cm2 | 62.2 ± 14.5 MΩ/cm2 | 9.8 ± 2.4 GΩ/cm2 |
4. Conclusions
We presented a technique that patterns cells on arrays of gold electrodes through ligand-mediated cell adhesion, and demonstrated that cell adhesion and spreading significantly influences the impedance characteristics of cell-based sensors. It is established through comparison studies that a tight cell binding on the electrode surface and full cell spreading to cover the surface area substantially improves impedance characteristics of cell-based sensors, which enables the detection of small cellular changes in response to external stimuli such as presence of drugs or toxins. The study suggests that patterning single cells on electrodes with dimensions as small as cell size through the mediation of covalently-bound short peptides would result in optimal cell binding and detection sensitivity. Single cell-based sensors allow for fundamental studies of cell biology without interference of cell-cell interactions, require minimal sample volumes for analyte detection, and offer rapid statistical analyses, high throughput data acquisition and portability.
Supplementary Material
Acknowledgments
The authors acknowledge the funding support from the National Institutes of Health (NIH/GMS) for the project of “Microelectrode arrays of single cell biosensors” (Grant R01GM075095) and lab assistance of Xinli Hu, Johnson Tey, and Kandy Yeung.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arndt S, Seebach J, Psathaki K, Galla HJ, Wegener J. Biosensors & Bioelectronics. 2004;19(6):583–594. doi: 10.1016/s0956-5663(03)00269-0. [DOI] [PubMed] [Google Scholar]
- Asphahani F, Zhang M. Analyst. 2007;132(9):835–841. doi: 10.1039/b704513a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceriotti L, Ponti J, Broggi F, Kob A, Drechsler S, Thedinga E, Colpo P, Sabbioni E, Ehret R, Rossi F. Sensors and Actuators B-Chemical. 2007;123(2):769–778. [Google Scholar]
- Fromherz P. European Biophysics Journal with Biophysics Letters. 1999;28(3):254–258. doi: 10.1007/s002490050206. [DOI] [PubMed] [Google Scholar]
- Giaever I, Keese CR. Proceedings Of The National Academy Of Sciences Of The United States Of America. 1991;88(17):7896–7900. doi: 10.1073/pnas.88.17.7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist KH, Giovangrandi L, Whittington RH, Kovacs GT. Biosensors and Bioelectronics. 2005;20(7):1397–1406. doi: 10.1016/j.bios.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Greve DW, Huang X, Nguyen D, Domach MM. Proceedings Of The Ieee. 2003;2(22–24):1358–1363. [Google Scholar]
- Hartmann C, Zozulya A, Wegener J, Galla HJ. Experimental Cell Research. 2007;313(7):1318–1325. doi: 10.1016/j.yexcr.2007.01.024. [DOI] [PubMed] [Google Scholar]
- Huang XQ, Nguyen D, Greve DW, Domach MM. Ieee Sensors Journal. 2004;4(5):576–583. [Google Scholar]
- Hynes RO. Cell. 1992;69(1):11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- Kataoka N, Iwaki K, Hashimoto K, Mochizuki S, Ogasawara Y, Sato M, Tsujioka K, Kajiya F. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(24):15638–15643. doi: 10.1073/pnas.242590799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs GTA. Proceedings Of The Ieee. 2003;91(6):915–929. [Google Scholar]
- Lo CM, Keese CR, Giaever I. Biophysical Journal. 1995;69(6):2800–2807. doi: 10.1016/S0006-3495(95)80153-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luong JHT. Analytical Letters. 2003;36(15):3147–3164. [Google Scholar]
- McFadden P. Science. 2002;297(5589):2075–2076. doi: 10.1126/science.297.5589.2075. [DOI] [PubMed] [Google Scholar]
- Mossop BJ, Barr RC, Zaharoff DA, Yuan F. Ieee Transactions on Nanobioscience. 2004;3(3):225–231. doi: 10.1109/tnb.2004.833703. [DOI] [PubMed] [Google Scholar]
- Omori T, Aonishi T, Miyakawa H, Inoue M, Okada M. Brain Research. 2006;1125:199–208. doi: 10.1016/j.brainres.2006.09.095. [DOI] [PubMed] [Google Scholar]
- Stenger DA, Gross GW, Keefer EW, Shaffer KM, Andreadis JD, Ma W, Pancrazio JJ. Trends In Biotechnology. 2001;19(8):304–309. doi: 10.1016/s0167-7799(01)01690-0. [DOI] [PubMed] [Google Scholar]
- Stett A, Egert U, Guenther E, Hofmann F, Meyer T, Nisch W, Haemmerle H. Anal Bioanal Chem. 2003;377(3):486–495. doi: 10.1007/s00216-003-2149-x. [DOI] [PubMed] [Google Scholar]
- Veiseh M, Veiseh O, Martin MC, Asphahani F, Zhang MQ. Langmuir. 2007;23(8):4472–4479. doi: 10.1021/la062849k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veiseh M, Wickes BT, Castner DG, Zhang MQ. Biomaterials. 2004;25(16):3315–3324. doi: 10.1016/j.biomaterials.2003.10.014. [DOI] [PubMed] [Google Scholar]
- Veiseh M, Zareie MH, Zhang MQ. Langmuir. 2002;18(17):6671–6678. [Google Scholar]
- Veiseh M, Zhang MQ. Journal Of The American Chemical Society. 2006;128(4):1197–1203. doi: 10.1021/ja055473q. [DOI] [PubMed] [Google Scholar]
- Xiao C, Luong JHT. Biotechnology Progress. 2003;19(3):1000–1005. doi: 10.1021/bp025733x. [DOI] [PubMed] [Google Scholar]
- Yin HY, Wang FL, Wang AL, Cheng J, Zhou YX. Analytical Letters. 2007;40(1):85–94. [Google Scholar]
- Ziegler C. Fresenius Journal of Analytical Chemistry. 2000;366(6–7):552–559. doi: 10.1007/s002160051550. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.