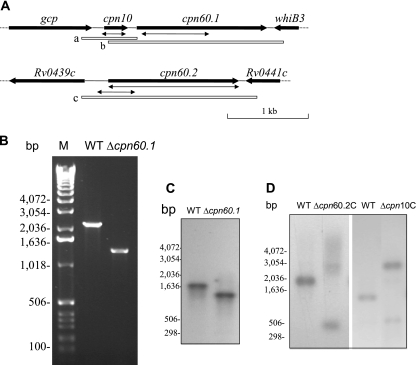
FIG. 1.
Construction of M. tuberculosis cpn60.1 mutant. (A) Genomic context of cpn10, cpn60.1, and cpn60.2. The double arrows indicate the deletions made in these genes. The open boxes a, b, and c show the locations of the fragments used in complementation experiments. (B) PCR confirmation of the deletion of the cpn60.1 gene, using the primers cpn60.1CF and cpn60.1CR. (C) Southern blotting analysis confirming the deletion of the M. tuberculosis H37Rv cpn60.1 gene. DNAs from the WT and the YHcpn60.1Δ strain were digested with NcoI and hybridized with probe b, shown in panel A. (D) Southern blotting analysis confirming that deletion of the M. tuberculosis H37Rv cpn60.2 or cpn10 gene was possible only when a WT copy of the gene was present. DNAs from the WT and the YHΔcpn60.2C strain were digested with AclI and Esp3I and hybridized with probe c, shown in panel A. Also, DNAs from the WT and the YHΔcpn10C strain were digested with PvuII and hybridized with probe a, shown in panel A. Since the WT copy of cpn60.2 or cpn10 was integrated into the chromosomal DNA, digestion with the restriction enzymes resulted in a larger fragment containing the WT copy of the gene. M, molecular weight marker (Invitrogen). The experiments were repeated twice, with identical results.