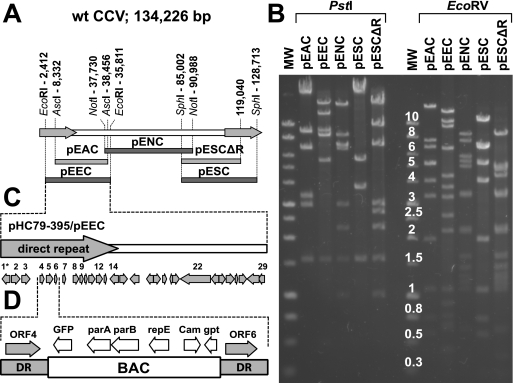
FIG. 1.
Overlapping CCV BACs. (A) Positions of the CCV fragments that were used to generate overlapping CCV BAC clones. The CCV genome (134.2 kb) contains two identical terminal direct repeats (DR) (18.6 kb each) (arrows) that flank the unique region (97.1 kb). The CCV restriction fragments that were used to generate overlapping CCV BACs are shown below the CCV genome and are aligned with the regions of the CCV genome from which they were derived. pEEC contains the EcoRI fragment (positions 2412 to 35811), pEAC contains the AscI fragment (positions 8332 to 38456), pENC contains the NotI fragment (positions 7730 to 90988), and pESC contains the SphI fragment (positions 85002 to 128713). Nucleotide positions correspond to the wt CCV genome. pESCΔR is a derivative of pESC from which the majority of direct terminal repeat was removed. pESCΔR contains only the first 3,403 bp of the right repeat region. (B) Restriction enzyme analysis of CCV BACs. pEEC, pENC, pESC, pEAC, and pESCΔR were digested with PstI or EcoRV, and restriction DNA fragments were separated on a 0.7% agarose gel. Restriction patterns of CCV BACs correspond to the predicted ones. MW, molecular weight marker with band size in kb (1-kb DNA ladder; Promega Corp.). (C) CCV sequences carried by pEEC and pHC79-395. These clones contain a part of the direct terminal repeat and a portion of the unique region of the CCV genome. Arrows indicate positions and orientations of the predicted CCV ORFs. 1* is a 5′-terminal part of CCV ORF1. CCV ORF12 carried by pEEC was deleted to produce pΔ12. (D) CCV ORF5 carried by pHC79-395 was replaced with the BAC cassette of plasmid pHA2 (1). parA, parB, and repE are regulatory genes derived from the F factor of E. coli. GFP, green fluorescent protein; Cam, chloramphenicol resistance; gpt, guanosine phosphoribosyl transferase.