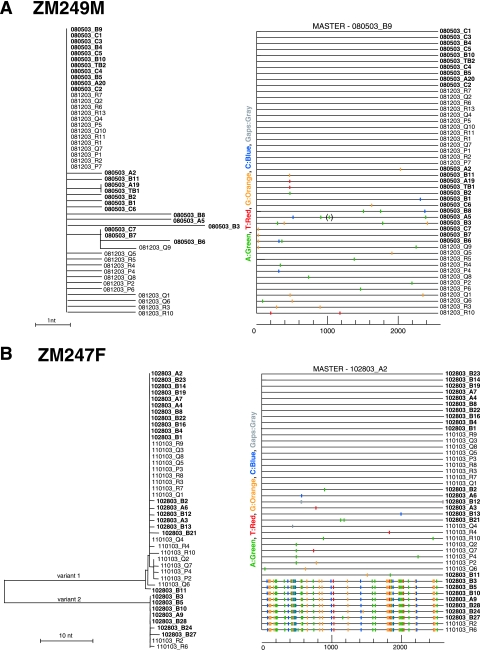
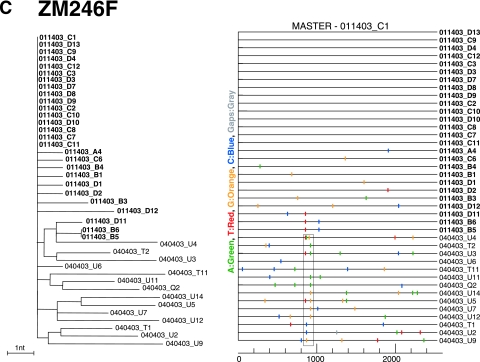
FIG. 3.
Identification of transmitted or early founder env genes in subjects with acute HIV-1 infection. SGA-derived env sequences from pre- and postseroconversion plasma samples from subjects ZM249M (A), ZM247F (B), and ZM246F (C) were examined by phylogenetic tree construction (left panels) and Highlighter analysis (right panels). Sequences from the earlier time points are in bold face. Trees are midpoint rooted. The scale bars represent 1 (A and C) or 10 (B) nt substitutions per site. The corresponding Highlighter diagrams denote the locations of nucleotide sequence substitutions in each env sequence in comparison to a reference sequence listed at the top; the positions of these substitutions within the env sequence are indicated at the bottom. Nucleotide substitutions and gaps are color coded. For each subject, sets of identical and nearly identical env sequences form consensus sequences that coalesce to transmitted or early founder viral env sequences. Subjects ZM249M and ZM246F were infected by a single virus and ZM247F by two viruses, identified as variant 1 and variant 2 in the tree (panel B). The tick in parentheses in sequence 080503_A5 (panel A) indicates a single-nucleotide insertion. The boxed region in panel C indicates clustered mutations in V3. Interestingly, these changes were limited to position 8 (Ile to Thr, Arg, or Lys) and position 25 (Asp to Asn, Glu, or Gly).