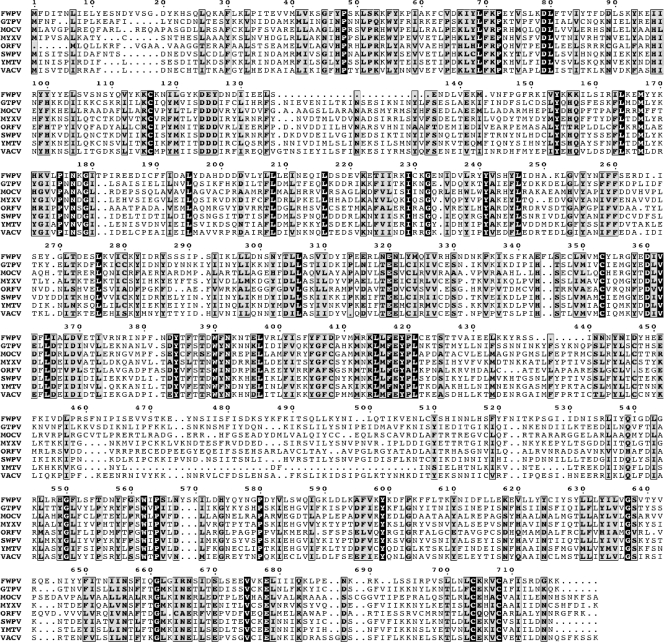
FIG. 1.
Multiple-sequence alignment of E2L orthologs. A multiple-sequence alignment was constructed with the T-Coffee algorithm (25). The alignment includes one representative amino acid sequence from each genus of Chordopoxvirus. Amino acids at invariant positions are shown as white letters on a black background; moderately conserved amino acids are shown as black letters against a gray background. Abbreviations: FWPV, fowlpox virus (Avipoxvirus); GTPV, goatpox virus (Capripoxvirus); MOCV, molluscum contagiosum (Molluscipoxvirus); MYXV, myxoma virus (Leporipoxvirus); ORFV, orf virus (Parapoxvirus); SWPV, swinepox virus (Suipoxvirus); YMTV, yaba monkey tumor virus (i); VACV, Orthopoxvirus.