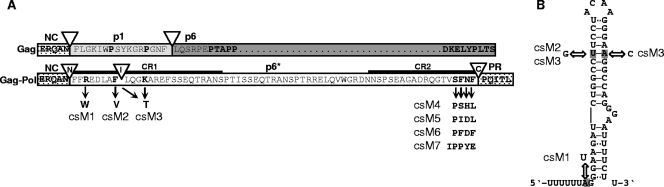
FIG. 1.
Mutagenesis of p6*-associated PR cleavage sites in the proviral context. (A) The entire HIV-1 BH10-derived p6* amino acid sequence (white box) is shown in the context of the Gag-Pol precursor with adjacent p7 and PR sequences (dotted boxes). The p6* region is overlapped by the gag open reading frame encoding the p1 (gray box) and p6 (dark gray box) proteins. PR cleavage sites are indicated by open triangles (N, amino-terminal; I, internal; C, carboxyl-terminal), and functional residues are depicted in boldface type. Amino acids replaced in p6* to alter PR cleavage sites are indicated by black arrows and are represented by provirus clones csM1 to csM7. CR1 and CR2 are conserved regions within p6*. (B) Proposed structure of the HIV-1 frameshift region according to Dulude et al. (13). Mutations introduced to modify amino-terminal (csM1) or internal (csM2, csM3) p6* cleavage are indicated by arrows. See text for more details.