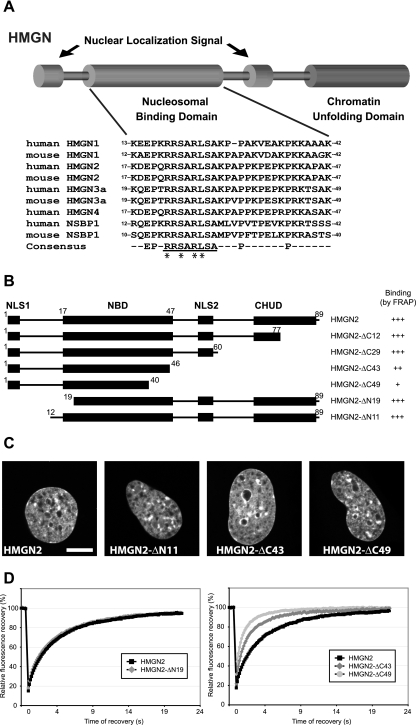
FIG. 1.
Mapping of HMGN2 NBD by FRAP on native chromatin in living cells. (A) Schematic representation of the structure of the HMGN proteins and alignment of the NBD of the mouse and human HMGNs. The numbers at the beginning of each sequence correspond to amino acid position of the first and last residues of the NBD in its respective sequence. The conserved HMGN motif within the consensus sequence is underlined. The amino acid residues that are critical for specific binding to nucleosomes are marked by asterisks (see the text). (B) Outline of the HMGN2 deletion mutants used to map the NBD in living cells. The known functional domains of the protein are indicated by black boxes (NLS, nuclear localization signal; CHUD, chromatin unfolding domain). For FRAP experiments, the proteins were fused to GFP at their C terminus. The name and the binding properties of each mutant are indicated on the right. Binding is rated from “+” (poor binding) to “+++” for wild-type binding (relative values in Table 1). (C) Intranuclear localization of the HMGN2 deletion mutants. Confocal images of live cells expressing the indicated GFP fusion proteins are shown. Scale bar, 10 μm. (D) Quantitative FRAP analysis of N-terminal (left) or C-terminal (right) deletion mutant expressed in MEFs.