Many inside and outside of the infectious disease community were surprised by a recent report from the Centers for Disease Control and Prevention that in 2005 (and presumably since then), methicillin-resistant Staphylococcus aureus (MRSA) caused more deaths in the United States than the human immunodeficiency virus/AIDS (2, 15). MRSA also caused more invasive infection than Streptococcus pneumoniae, Streptococcus pyogenes, Neisseria meningitidis, and Haemophilus influenzae (15). It is important to note that these numbers represent only S. aureus infections confounded by methicillin resistance, with more than twice as many infections caused by S. aureus strains that have not yet acquired this level of resistance. As a leading threat to public health, there is urgency to the development of new therapeutic and preventive strategies for S. aureus infection. In this issue of the Journal of Bacteriology, Majerczyk et al. (16) report that CodY, a global regulator that senses the nutritional status of the bacterium, contributes to the decision by S. aureus of “when and in what amounts” to express virulence. This observation connects sensing of the nutritional surroundings to regulation of virulence by this leading cause of invasive human infection.
Based on limited sampling areas, such as those probed by nasal swab, S. aureus carriage rates are usually found to be 33 to 50% among healthy study populations (see, for example, references 22 and 31). This implies that if the entire host were queried, most if not all humans would be found to be colonized by S. aureus, defining the organism as a human commensal. Despite its existence as a commensal, or perhaps because of the intimate association accorded by this relationship, S. aureus has emerged as a leading cause of infection in hospitals, as well as in the general community. Compounding this problem is the emergence and spread of antibiotic resistance, including resistance to penicillinase stable β-lactams, such as oxacillin and methicillin (giving rise to the nomenclature for MRSA) (9, 14), and more recently resistance to the last-line antibiotic, vancomycin (4). Although a rarity just 10 years ago (3, 13) fully one-third of MRSA infections now occur in the community (15). Community-acquired MRSA infections most commonly begin as an infection of the skin or soft tissues, with a fraction progressing to severe invasive infection, bacteremia, and death (3, 5, 11).
In the United States it is believed that the current outbreak of invasive community-acquired MRSA infection was triggered by acquisition by a community S. aureus strain of the methicillin resistance-conferring cassette (staphylococcal cassette chromosome mec [or SCCmec]) (9, 14) and a second determinant, possibly from skin-colonizing S. epidermidis, encoding genes for the catabolism of arginine, known as the arginine catabolic mobile element (or the ACME locus) (10). Together, these changes resulted in the emergence in the community of a virulent MRSA lineage termed USA300. Infection-derived isolates such as USA300 express more than 30 toxins and adhesins believed to contribute to S. aureus virulence, including Panton-Valentine leukocidin; alpha-, beta-, delta-, and γ gamma-hemolysins; and more than 20 enterotoxins and superantigens (1, 10).
A question central to understanding the biology of the S. aureus-human dynamic is why would a commensal express such a plethora of toxins? A well-reasoned argument was recently advanced that suggests that the toxins facilitate transmission (19). It was also considered that these toxins may promote the colonization of the wet mucosa, which, unlike the keratinized skin, is actively defended by phagocytic cells of the innate immune system (19). A third possibility is that toxigenic commensals, such as S. aureus, evolved complex regulatory schemes to prevent causing infection in a healthy host; however, at the first sign of weakness, such commensals are perfectly positioned to hasten the demise of the host and be first to the table. The virtue of this perspective is that in the traditional view of a balanced predator-prey relationship, there is little long-term negative consequence to either the predator or the prey in culling weakened individuals from the herd, and there is considerable advantage to the predator in being the first to the table in a highly competitive environment.
The mechanisms by which the expression of toxins, adhesins, and biofilm formation are regulated by S. aureus are complex and not completely understood. One level of regulation is achieved by the autoinduction, quorum-sensing accessory gene regulator, or agr, system (1) (Fig. 1). A second level of global regulation is achieved by a family of winged helix DNA-binding proteins related to SarA (6). In this issue, Sonenshein and coworkers show that there is a third level of regulation provided by the nutritionally controlled regulator, CodY (16).
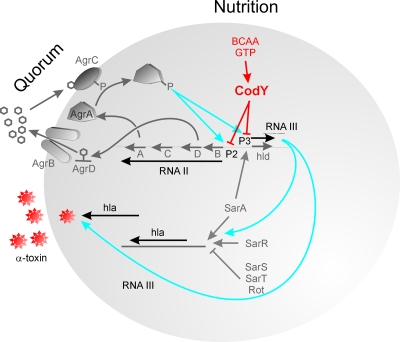
FIG. 1.
Factors influencing the control of virulence of S. aureus include quorum regulation and now the nutritional quality of the environment (for reviews and more details, see references 21 and 30). Extracellular accumulation of the AgrD-derived thiolactone-containing peptide provides a mechanism for quorum regulation of virulence. However, as shown in the study by Majerczyk et al. (16), CodY influences the expression of RNAII and RNAIII from P2 and P3, respectively, limiting in the presence of BCAA and GTP the quorum activator of virulence and its effector, RNAIII. RNAIII, when made in response to quorum accumulation or when not repressed by CodY (such as in codY mutants or parental cells with low intracellular pools of GTP and BCAA), enhances both transcription and translation of alpha-toxin and other S. aureus toxins and exoproteins and also PIA/PNAG-associated biofilm formation.
The staphylococcal agr quorum-sensing system, when a quorum is reached, represses expression of several cell surface proteins and adhesins and enhances the expression of many toxins. This quorum is achieved in vitro during the transition from the late-exponential phase to the stationary phase (21). Two divergent transcripts, RNAII and RNAIII, are generated at the agr locus, originating from promoters P2 and P3, respectively (Fig. 1). The P2 transcript encodes four proteins that constitute the agr-sensing mechanism. AgrB is a transmembrane protein that appears to be involved in (i) proteolytic trimming of agrD into an oligopeptide; (ii) modification of the peptide by the formation of a cyclic thiolactone bond between an internal cysteine and the carboxyl terminus; and (iii) secretion of the autoinducing peptide (AIP) quorum signal (21). AgrA and AgrC form a two-component regulatory system in which the sensor kinase, AgrC, binds extracellular AIP (Fig. 1). Subsequent phosphorylation activates the AgrA response regulator. Through an as-yet-undefined mechanism, phosphorylated AgrA leads to increased P2 and P3 transcription. Sequence variation in agrB, agrD, and agrC has led to the evolution of at least four agr specificity groups, in which AIP produced by one group inhibits the function of other agr types (21). Increased transcription from P3 not only increases levels of message for δ-hemolysin (hld) included within RNAIII but, as shown in Fig. 1, RNAIII significantly increases both the transcription and the translation of secreted virulence factors, including alpha-hemolysin and other toxins (30).
Staphylococcal accessory regulator (SarA) is a DNA-binding protein that appears to be required for full agr transcription (7). SarA affects the expression of a wide array of virulence genes, in some cases independently of agr. SarA is responsive to some environmental conditions through intermediate regulators (8) and likely affects the expression of the agr locus accordingly. SaeRS (not shown) is a two-component system with a global effect on regulation of virulence factors. sae mutants produce substantially less hemolysin and coagulase but do not appear to affect the production of RNAIII. The expression of at least one sae transcript was decreased in an agr mutant, suggesting that the sae system acts downstream of agr (12).
In this issue of the Journal of Bacteriology, Majerczyk and coworkers (16) report a functional connection between CodY and virulence gene expression by S. aureus. CodY, together with CcpA and TnrA, control the flow of carbon and nitrogen in low G+C gram-positive bacteria (29). CodY is a highly conserved DNA-binding protein that regulates gene expression in response to nutritional status, by interacting with either of two intracellular metabolites: GTP or branched-chain amino acids (BCAA) (24, 27, 28). In the presence of elevated intracellular concentrations of these cofactors, CodY is activated and binds to DNA consensus sequences, known as the “CodY box.” In Bacillus subtilis, this binding regulates the expression of many carbon and nitrogen metabolism genes, transport genes, and sporulation and competence genes (20, 26).
Malke et al. (17, 18) observed that CodY influenced virulence gene expression in the human pathogen, S. pyogenes. CodY was observed to control genes related to nitrogen metabolism, including extracellular protein-degrading enzymes PrtS, ScpA, and IdeS (17). However, CodY was also found to influence global regulators of virulence and other transcription factors including pel/sagA, mga, covRS, ropB, and pyrR. By controlling the expression of other regulators, the effect of CodY sensing of nutritional status is amplified and extends to many aspects of pathogen behavior. CodY was observed to be functional in regulating S. pyogenes virulence gene expression in blood (18), indicating that at least for this bacterium, blood allows accumulation of sufficient intracellular pools of BCAA and GTP to activate CodY.
Because little was known about a possible connection between nutritional status and virulence for S. aureus, Majerczyk et al. (16) investigated the role of CodY by using two clinical isolates, UAMS-1 and SA564. codY-null mutants were generated by allelic replacement, and these mutants were subjected to a variety of tests including hemolysin assays, detection of alpha-toxin by Western blotting, detection of polysaccharide intercellular adhesin (PIA/PNAG, encoded by the icaR regulated icaADBC operon) by immunoblotting, and determination of biofilm formation. Transcriptional analysis of icaA, icaR, RNAII, and RNAIII were performed by reverse transcription-PCR during the exponential and stationary phases (16). Mutants defective in codY expression formed more biofilm than parental strains and produced more PIA. This indicates that for wild-type S. aureus cultured in broth, CodY limits the expression of the icaADBC operon independently of the repressor icaR (16). Mutant strains defective in codY expression produced higher levels of hemolytic activity in the stationary phase, and the alpha-toxin gene (hla) was observed to be expressed at higher levels, indicating that increased hemolytic activity was attributable to derepression in the codY mutant (16).
Importantly for global regulation of virulence, mutation in codY led to increased transcription from both P2 and P3 promoters of the accessory gene regulator (agr) locus in the exponential phase of growth. If CodY functions in S. aureus identically to what has been observed in other organisms, this indicates that in a nutritionally rich environment, such as blood, CodY serves to dampen the expression of key virulence traits, including biofilm formation, alpha-toxin expression, and global virulence expression as regulated by agr.
Assuming all of this to be reflective of conditions in infection (it has yet to be directly determined in invasive infection whether CodY is actually active or inactive and whether its effects on virulence factor expression in that environment are overridden by others factors currently known or unknown), why would a bacterium possess virulence traits that it downregulates in invasive infection? It may be that these virulence traits are in fact colonization factors that enable the bacterium to stably inhabit the well-defended mucosal surface in a harmless, homeostatic way. Perhaps in the mucosa, CodY does not repress exoprotein expression, and these “virulence traits” hold mucosal defenses at bay, but not more. Perhaps this highly coevolved commensal has learned to restrict its virulence in the noncommensal niche. In 2007 Palmer et al. in the Whiteley laboratory (23) characterized the nutritional environment in sputum and derived an artificial medium to examine Pseudomonas aeruginosa gene expression in that environment. Similar experiments comparing gene expression by codY mutants and parental S. aureus strains in a mucosa-like environment would be highly informative in helping to elucidate S. aureus behavior in natural colonization. If the regulation of CodY activity is central to mucosal colonization, it may be possible to develop new methods to influence colonization (i.e., reduce MRSA colonization in health care workers) by supplementing the availability of nontoxic BCAA in the mucosa.
Although CodY suppresses virulence trait expression, especially in exponential growth, once S. aureus cells are abundant, the virulent phenotype is expressed through activation of the agr quorum system. Limiting virulence factor expression and biofilm formation in an environment such as blood at low bacterial cell densities would likely facilitate phagocytic clearance, perhaps perpetuating the stable relationship between host and commensal. However, if for some reason the host is unable to clear these low densities of cells, perhaps because of senescence of the immune system, serious coinfection, or other injury (or in the current day, hospitalization, diabetes, or other host stressor), S. aureus may reach a threshold that trips the unbridled expression of virulence, precipitating the demise of the host. Supporting the notion that S. aureus has evolved control systems that limit virulence in an immunologically competent host, it was observed previously by Gresham and coworkers (25) that the AgrD-derived thiolactone containing quorum signal is oxidized and inactivated by reactive oxygen and nitrogen species within phagocytic cells and that this in fact attenuates infection. In such a model, S. aureus behavior would parallel that of predators that depend on pack behavior for predation. Just as pack feeding culls weakened individuals from a herd, S. aureus may be constantly probing the human immune status waiting for a sign of weakness. Intriguing evidence for pack-like attack on damaged human cells by motile P. aeruginosa has been obtained using real-time video in the S. Fleiszig laboratory (http://vision.berkeley.edu/patliv/), so perhaps the comparison is apt.
Pathogen and commensal behaviors are clearly influenced by environmental cues, including the availability of select nutrients, such as BCAA. This is clear for mucosal pathogens, including S. pyogenes (17, 18), P. aeruginosa (23), and now S. aureus (16). Conceptually, it may be important to view sensitivities to nutrients, such as BCAA (and potentially vitamins and other auxotrophies as well), as important environmental signaling molecules. The more we understand how natural environments affect the behavior of bacteria that infect humans, the better we will be able to regulate the composition of these environments and thus the host-pathogen dynamic.
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
Footnotes
Published ahead of print on 25 January 2008.
REFERENCES
- 1.Arvidson, S. 2004. Virulence gene regulation and their role in pathogenesis of disease, p. 154-176. In D. Aldeen and K. Hiramatsu (ed.), Staphylococcus aureus: molecular and clinical aspects. Horwood Publishing, Chichester, United Kingdom.
- 2.Bancroft, E. A. 2007. Antimicrobial resistance: it's not just for hospitals. JAMA 2981803-1804. [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 1999. Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus—Minnesota and north Dakota, 1997-1999. MMWR Morb. Mortal. Wkly. Rep. 48707-710. [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 2002. Vancomycin-resistant Staphylococcus aureus—Pennsylvania, 2002. MMWR Morb. Mortal. Wkly. Rep. 51902. [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2006. Community-associated methicillin-resistant Staphylococcus aureus infection among healthy newborns—Chicago and Los Angeles County, 2004. MMWR Morb. Mortal. Wkly. Rep. 55329-332. [PubMed] [Google Scholar]
- 6.Cheung, A. L., K. A. Nishina, M. P. Trotonda, and S. Tamber. The SarA protein family of Staphylococcus aureus. Int. J. Biochem. Cell. Biol., in press. [DOI] [PMC free article] [PubMed]
- 7.Cheung, A. L., M. G. Bayer, and J. H. Heinrichs. 1997. sar genetic determinants necessary for transcription of RNAII and RNAIII in the agr locus of Staphylococcus aureus. J. Bacteriol. 1793963-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deora, R., T. Tseng, and T. Misra. 1997. Alternative transcription factor sSB of Staphylococcus aureus: characterization and role in transcription of the global regulatory locus sar. J. Bacteriol. 1796355-6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deurenberg, R. H., C. Vink, S. Kalenic, A. W. Friedrich, C. A. Bruggeman, and E. E. Stobberingh. 2007. The molecular evolution of methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 13222-235. [DOI] [PubMed] [Google Scholar]
- 10.Diep, B., S. Gill, R. Chang, T. Phan, J. Chen, M. Davidson, F. Lin, J. Lin, H. Carleton, and E. Mongodin. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired methicillin-resistant Staphylococcus aureus. Lancet 367731-739. [DOI] [PubMed] [Google Scholar]
- 11.Fridkin, S. K., J. C. Hageman, M. Morrison, L. T. Sanza, K. Como-Sabetti, J. A. Jernigan, K. Harriman, L. H. Harrison, R. Lynfield, and M. M. Farley. 2005. Methicillin-resistant Staphylococcus aureus disease in three communities. N. Engl. J. Med. 3521436-1444. [DOI] [PubMed] [Google Scholar]
- 12.Giraudo, A. T., C. Mansilla, A. Chan, C. Raspanti, and R. Nagel. 2003. Studies on the expression of regulatory locus sae in Staphylococcus aureus. Curr. Microbiol. 46246-250. [DOI] [PubMed] [Google Scholar]
- 13.Herold, B. C., L. C. Immergluck, M. C. Maranan, D. S. Lauderdale, R. E. Gaskin, S. Boyle-Vavra, C. D. Leitch, and R. S. Daum. 1998. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA 279593-598. [DOI] [PubMed] [Google Scholar]
- 14.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types pf staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 451323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klevens, R. M., M. A. Morrison, J. Nadle, S. Petit, K. Gershman, S. Ray, L. H. Harrison, R. Lynfield, G. Dumyati, J. M. Townes, A. S. Craig, E. R. Zell, G. E. Fosheim, L. K. McDougal, R. B. Carey, S. K. Fridkin, et al. 2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 2981763-1771. [DOI] [PubMed] [Google Scholar]
- 16.Majerczyk, C. D., M. R. Sadykov, T. T. Luong, C. Lee, G. A. Somerville, and A. L. Sonenshein. 2008. Staphylococcus aureus CodY represses virulence gene expression. J. Bacteriol. 1902257-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malke, H., K. Steiner, W. M. McShan, and J. J. Ferretti. 2006. Linking the nutritional status of Streptococcus pyogenes to alteration of transcriptional gene expression: the action of CodY and RelA. Int. J. Med. Microbiol. 296259-275. [DOI] [PubMed] [Google Scholar]
- 18.Malke, H., and J. J. Ferretti. 2008. CodY-affected transcriptional gene expression of Streptococcus pyogenes during growth in human blood. J. Med. Microbiol. 56707-714. [DOI] [PubMed] [Google Scholar]
- 19.Massey, R. C., M. J. Horsburgh, G. Lina, M. Höök, and M. Recker. 2006. The evolution and maintenance of virulence in Staphylococcus aureus: a role for host-to-host transmission? Nat. Rev. Microbiol. 4953-958. [DOI] [PubMed] [Google Scholar]
- 20.Molle, V., Y. Nakaura, R. P. Shivers, H. Yamaguchi, R. Losick, Y. Fujita, and A. L. Sonenshein. 2003. Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis. J. Bacteriol. 1851911-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 481429-1449. [DOI] [PubMed] [Google Scholar]
- 22.Ohara-Nemoto, Y., H. Haraga, S. Kimura, and T. K. Nemoto. 2008. Occurrence of staphylococci in the oral cavities of healthy adults and nasal oral trafficking of the bacteria. J. Med. Microbiol. 5795-99. [DOI] [PubMed] [Google Scholar]
- 23.Palmer, K. L., L. M. Aye, and M. Whiteley. 2007. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J. Bacteriol. 1898079-8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ratnayake-Lecamwasam, M., P. Serror, K. W. Wong, and A. L. Sonenshein. 2001. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev. 151093-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rothfork, J. M., G. S. Timmins, M. N. Harris, X. Chen, A. J. Lusis, M. Otto, A. L. Cheung, and H. D. Gresham. 2004. Inactivation of a bacterial virulence pheromone by phagocyte-derived oxidants: new role for the NADPH oxidase in host defense. Proc. Natl. Acad. Sci. USA 10113867-13872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Serror, P., and A. L. Sonenshein. 1996. CodY is required for nutritional repression of Bacillus subtilis genetic competence. J. Bacteriol. 1785910-5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shivers, R. P., and A. L. Sonenshein. 2004. Activation of the Bacillus subtilis global regulator CodY by direct iteration with branched-chain amino acids. Mol. Microbiol. 53599-611. [DOI] [PubMed] [Google Scholar]
- 28.Slack, F. J., P. Serror, E. Joyce, and A. L. Sonenshein. 1995. A gene required for nutritional repression of the Bacillus subtilis dipeptide permease operon. Mol. Microbiol. 15689-702. [DOI] [PubMed] [Google Scholar]
- 29.Sonenshein, A. L. 2007. Control of key metabolic intersections in Bacillus subtilis. Nat. Rev. Microbiol. 5917-927. [DOI] [PubMed] [Google Scholar]
- 30.Yarwood, J. M., and P. M. Schlievert. 2003. Quorum sensing in Staphylococcus infections. J. Clin. Investig. 1121620-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zafar, U., L. B. Johnson, M. Hanna, K. Riederer, M. Sharma, M. G. Fakih, M. C. Thirumoorthi, R. Farjo, and R. Khatib. 2007. Prevalence of nasal colonization among patients with community-associated methicillin-resistant Staphylococcus aureus infection and their household contacts. Infect. Control Hosp. Epidemiol. 28966-969. [DOI] [PubMed] [Google Scholar]