Fig. 4.
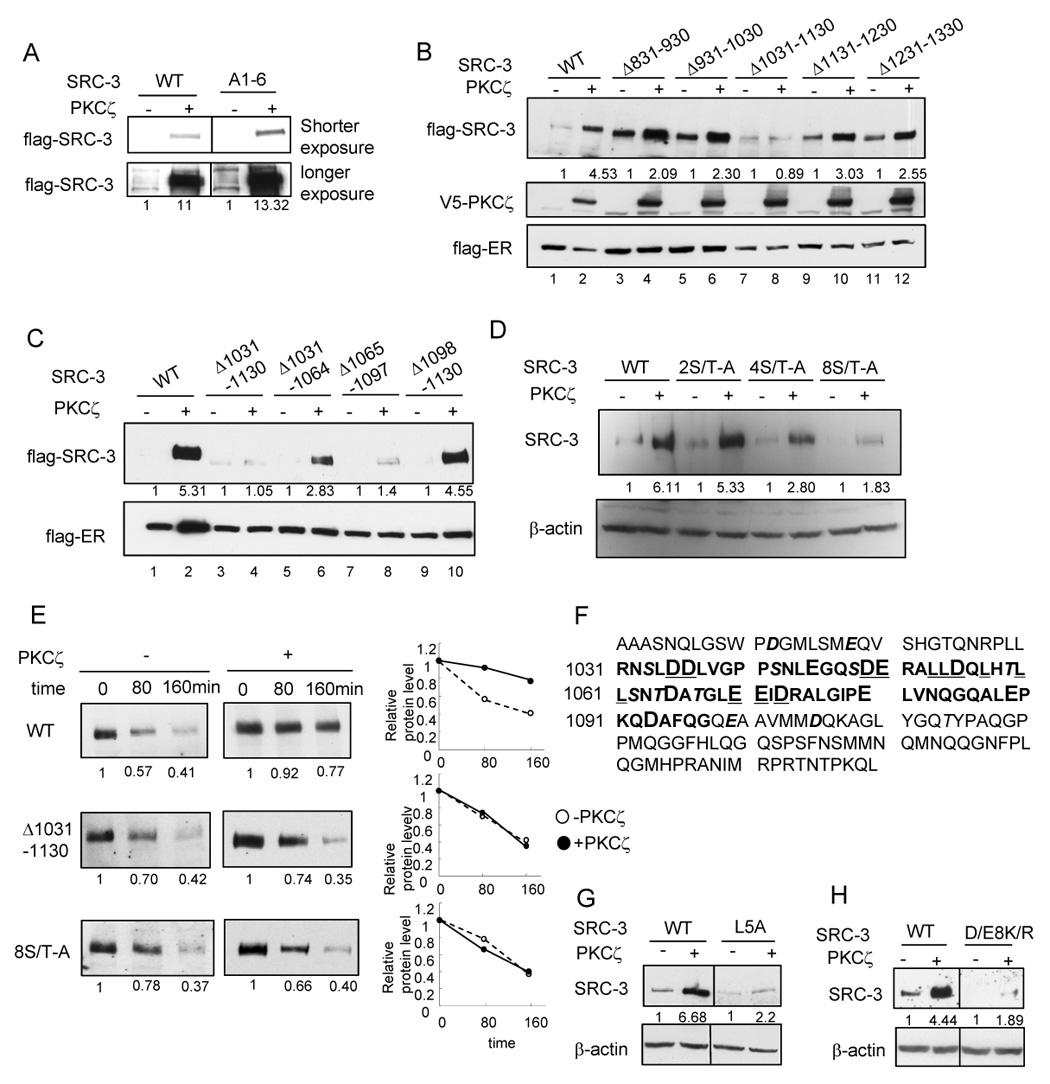
Identification of an important region in SRC-3 for aPKC stabilization effect (A) Effects of aPKC on SRC-3 A1-6 mutant (B) Effects of aPKC on SRC-3 deletion mutants (C) Effects of aPKC on SRC-3 mutants with deletion within the region of 1031–1130 (D) Effects of aPKC on SRC-3 mutants with Ser/Thr-Ala mutations within the region of 1031–1130. Details of the mutants were described in Supplemental Procedures. (E) Protein stability of SRC-3 wild type and the mutants as assayed by treating cells with 200 µg/ml cycloheximide at indicated time period. Shown in the left panel is the Western blot analysis of flag-SRC-3 protein. The right panel shows the quantification of protein intensity. (F) Amino acid sequences of SRC-3 1031–1097 (bolded) and the surrounding sequences. Acidic residues are in larger fonts. Ser/Thr residues are in Italic and shadowed. (G) L5A mutation (shown underlined in panel A) significantly reduced aPKC stabilization effect. (H) D/E8K/R mutation (shown underlined in panel A) significantly reduced aPKC stabilization effect.
