Figure 4. Distinct hippocampal gene expression profiles are associated with distinct stages of learning & memory.
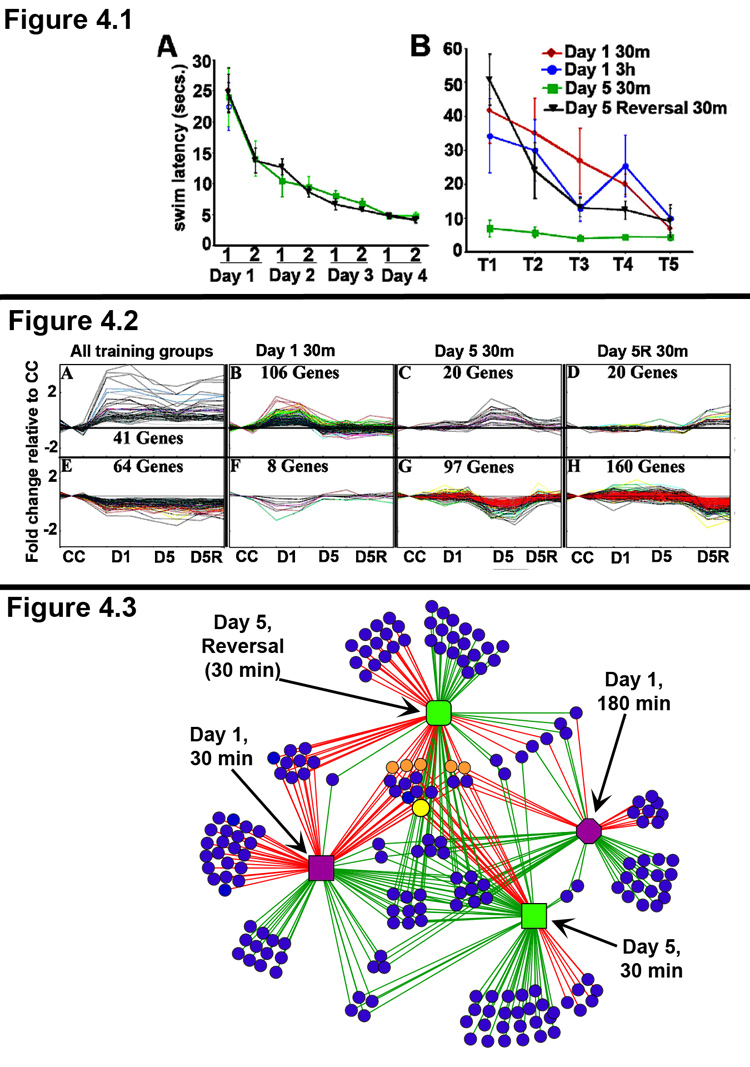
(Panel 4.1) Behavioral performance in the spatial water maze task. Different groups of rats were trained in the spatial water maze task, as described in the text. N = 6 rats per group. Panel A: The rats given multiple days of training learned the task well and showed an asymptotic performance achieved by end of day 3. Panel B: The behavioral performance of each group in the last training session before being sacrificed is shown. Note, that the Day 5 reversal rats were required to extinguish their response to the old platform location, in order to locate the new platform location. Accordingly, the behavioral performance of the Day 5 Reversal rats is similar to that of the Day 1 rats.
(Panel 4.2) Pattern template matching of microarray expression data reveals gene expression profiles associated with distinct stages of learning. RNA was isolated from the dorsal hippocampi of 6 rats from each behavioral group, and RNA from 3 rats each was used to generate replicate pools for each behavioral condition. Genome-wide gene expression analysis was done using Affymetrix® GeneChip® Rat230_2 arrays for each replicate pool. The Affymetrix PLIER algorithm was used to generate normalized gene expression values. TIGR MeV was used to define a set of ~400 significant genes with an ANOVA value of <0.05 and a fold change of < 1.5 or >1.5 relative to the caged control value. The gene expression profiles shown here were generated using a pattern template matching clustering algorithm. Gene expression levels are normalized to caged controls (CC), and the group labels are listed at the bottom of the panel (CC, D1, D5, and D5R). The data for each replicate chip is shown as a single data point, resulting in the “mountains” (panels B, C, and D) or “valleys” (panels F, G, and H) for genes differentially expressed in only one behavioral condition. Panels A & E show several genes that are induced (A) and repressed (E) in all behavior groups at 30m post-behavior. Panels B–D and F–H show distinct clusters of genes that are regulated only by initial learning (B &F), overtraining (reference memory retrieval; C &G), or reversal learning (reference memory extinction, coincident with new information acquisition; D &H). Notably, although the behavioral performance of D1 and D5R rats were similar, the gene expression profiles are distinct. Additionally, several genes in the D5 30m expression profile are associated with axonal outgrowth and new synapse formation, suggesting that overtraining might recruit additional plasticity mechanisms which play a role in the long-term stability of highly learned information (Reckart et al., 2007).
(Panel 4.3) Complex network analysis of gene expression networks regulated by distinct stages of learning & memory. Network of gene expression regulated by behavior in the spatial water maze task at D1 30m (purple square), D1 3h (purple octagon), D5 30m (green square), D5R 30m (green rounded-square). The lines connect behavior groups with the genes (blue circle nodes) that are differentially regulated by that behavior. Red & Green edges indicate up- and down- regulation of gene expression, respectively. Note that the several nodes in the center of the network represent genes that are regulated across multiple stages of learning and memory. This is also demonstrated by the high connectivity of these nodes in the network. Of these “core” genes several common IEGs are indicated as orange circle nodes and Arc is shown as a larger yellow circle node. In contrast, low connectivity genes, represented by blue circles connected to only one behavior group, are regulated only by a single behavior (i.e., in a distinct state of learning and memory). The degree of similarity or difference of the gene expression networks between any two of the behavior groups (stages of learning and memory) can be culled from the number of shared and distinct regulated genes. For example, D1 30m and D5R 30m exclusively share 9 up-regulated and 1 down regulated genes (left side of figure).
