Figure 5. Modification of the Plasticity Interactome by IEG Expression.
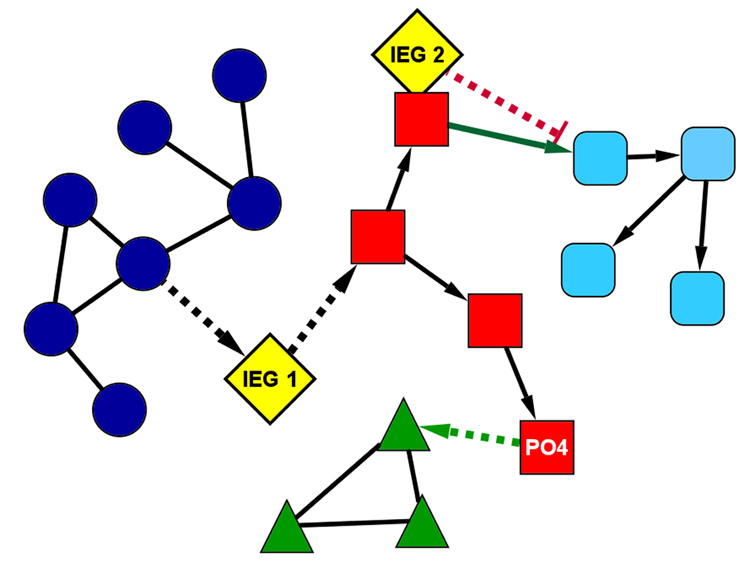
A simplified model of a protein interaction network containing 4 distinct subnetworks (dark blue circles, red squares, green triangles, and aqua rounded-squares). Solid lines between protein nodes represent constitutive interactions and dotted lines represent conditional interactions. Directionality of interactions is noted with arrowheads. Prior to patterned synaptic input, the two IEGs (IEG1 and IEG2, yellow diamonds) are absent and the protein in the red subnetwork is not phosphorylated. Under these basal conditions, the green and blue subnetworks function in isolation, and the red and aqua subnetworks interact. Following patterned synaptic activity, IEGs 1 and 2 are induced and the protein in the red network is phosphorylated (indicated by “PO4”), activating the conditional interactions. With these conditional interactions now active, the dialog between the subnetworks changes dramatically. IEG1 couples the blue and red subnetworks, and IEG2 functions as a dominant negative gene to block the interaction between red and aqua subnetworks. The phosphorylation in the red subnetwork now enables interaction with the green subnetwork. These 3 changes (induction of the two IEGs and the phosphorylation in the red subnetwork) markedly change the behavior of the entire network, with now a directed interaction between blue, red, and green networks, which did not occur in the basal state. Please note the powerful capacity of IEGs to modify the dialog of the plasticity interactome, and by regulation of a relatively limited number of IEGs.
