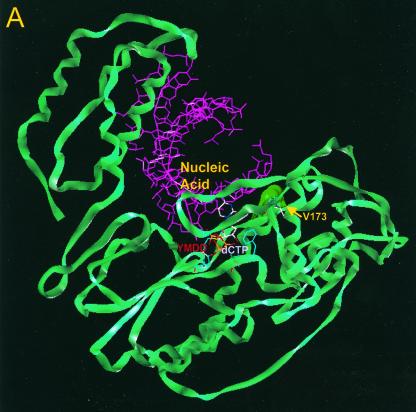
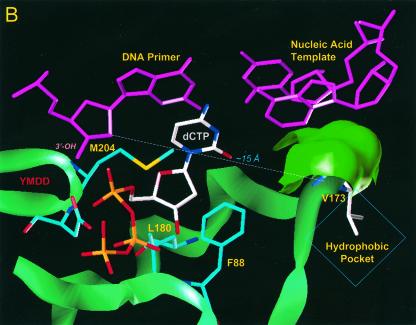
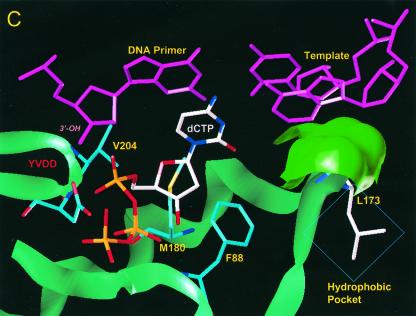
FIG. 4.
Location of residue rtV173 within HBV polymerase and modeling of the leucine substitution. (A) A molecular model of HBV polymerase based on the crystal structures of HIV type 1 RT and Moloney murine leukemia virus RT has been described elsewhere (12). The location of residue rtV173 and its molecular surface (indicated in bright green) are shown in the context of the entire enzyme. Nucleic acids representing the primer and template strands of DNA are indicated in magenta. The location of the YMDD motif is indicated. dCTP is docked in the nucleotide binding pocket of the polymerase. (B) Close-up view of the wild-type polymerase active site, showing the position of rtV173 beneath the template DNA strand. (C) Similar close-up view, modeling the rtV173L-rtL180M-rtM204V mutant active site. Note the position shift of residue F88.