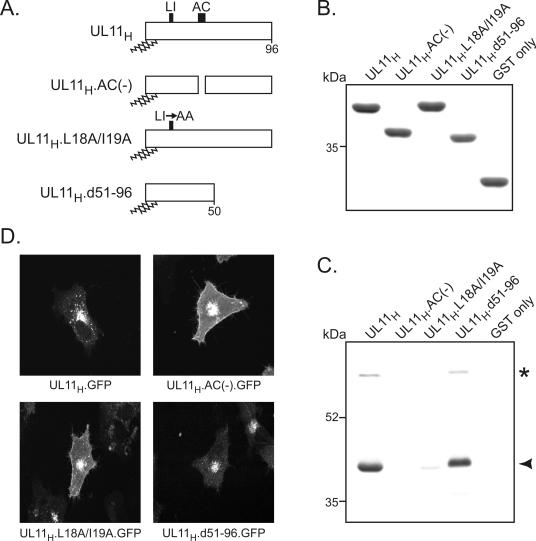
FIG. 2.
Identification of UL11-binding partners by a GST pull-down assay. (A) Diagrams of UL11 constructs. The 96-amino-acid UL11 sequence from HSV-1 is shown; the positions of the acidic cluster (AC) and the dileucine motif (LI) are indicated. N-terminal myristylation and palmitylation are denoted by wavy lines. (B) Coomassie blue-stained gel of GST fusion proteins. The UL11 constructs shown in panel A were fused to the C terminus of the GST protein and purified from Escherichia coli cells. Approximately equal amounts of proteins were used in all reactions. (C) Proteins pulled down by the GST-UL11 constructs. Purified GST fusion proteins were incubated with radiolabeled, HSV-1-infected cell lysates and washed with NP-40 lysis buffer. Proteins bound to the GST constructs were separated by SDS-10% PAGE, and radiolabeled proteins were detected by autoradiography. The asterisk and the arrowhead denote the 69- and 40-kDa proteins, respectively. (D) Subcellular localization. The UL11 constructs shown in panel A were fused to the N terminus of GFP, expressed in A7 cells, and visualized 18 h later by live-cell confocal microscopy.