Figure 1. Intrinsic Terminators and Single-Molecule DNA-pulling Assay.
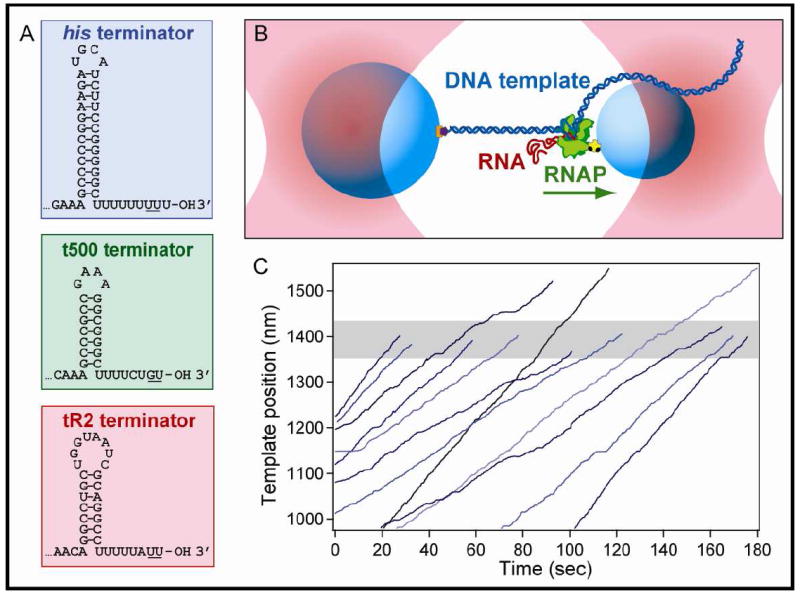
(A) Sequences and expected secondary structure for the three intrinsic hairpin terminators and U-tracts used in this study. Underlined bases indicate the transcript termination positions.
(B) Experimental geometry for the DNA-pulling dumbbell assay (not to scale). Two polystyrene beads (light blue) are held in two separate optical traps (pink). RNAP (green) is attached to the smaller of the two beads via a biotin-avidin linkage (yellow and black), and the DNA template (dark blue) is attached to the larger bead via a digoxigenin-antidigoxigenin linkage (purple and orange). The RNA transcript (red) emerges co-transcriptionally from RNAP and is untethered. The direction of transcription is shown (green arrow); in this particular orientation of the DNA template, the applied tension assists RNAP translocation.
(C) Twelve representative records of RNAP elongation on the his terminator template under 18 pN of assisting load. Nine records terminated within ±40 nm of the expected position of the terminator (shaded gray area), whereas three ran through the terminator position.
