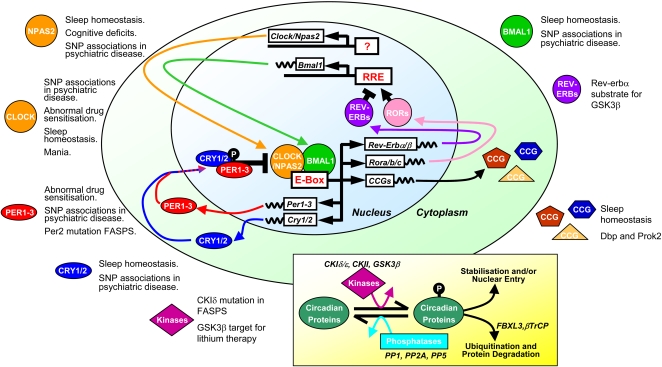
Figure 1. The Mammalian Molecular Circadian Oscillator.
The molecular circadian oscillator incorporates numerous transcriptional and posttranslational elements. Disruptions in many of the individual circadian elements in mice can lead to behavioural disturbances that mirror endophenotypes in human neurological and psychiatric disorders. Moreover, some studies have established circadian gene polymorphisms in psychiatric conditions and mutations in behavioural syndromes (see text for details). The central component of the figure depicts the core mammalian circadian feedback loop. CLOCK(or NPAS2):BMAL1 heterodimers drive the transcription of multiple genes (Cry1/2, Per1-3, Rev-Erba/b, Rora/b/c, multiple CCGs) through E-box elements. Nuclear accumulation of CRY and PER proteins can inhibit CLOCK:BMAL1-mediated transcription by directly interacting with the complex (black bar–ended arrow). As PER and CRY levels fall, the negative repression is lifted and CLOCK:BMAL1-driven transcription re-occurs. In an additional stabilising loop, REV-ERB and ROR proteins co-regulate the transcription of Bmal1 by competing for RREs in its promoter sequence. Rhythmic output of the clock is achieved through E-box elements in CCG which can impact a range of cell processes and physiology. The stability and subcellular localisation of circadian proteins is highly regulated by kinases and phosphatases (inset box). Although not entirely understood, the phosphorylation state of circadian proteins can affect their cellular localisation and/or stability. Mutations affecting the stability of Per proteins can accelerate the molecular clock in humans, leading to the inherited syndrome familial advanced sleep phase syndrome (FASPS).