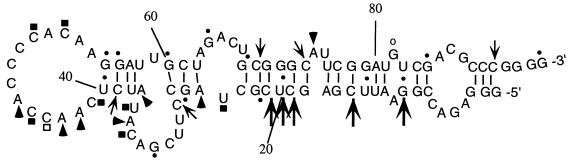
Figure 1.
Predicted secondary structure of R8-35 RNA. The computer-predicted secondary structure of the cap-binding RNA is shown. Susceptibilities of residues to various single- and double-stranded specific nucleases are indicated. Strong reactivities are marked by filled-in symbols and weak reactivities are denoted with open symbols. RNase A (□ and ▪) displays specificity for single-stranded U and C residues. RNase T1 (○ and •) cleaves 3′ of single-stranded G residues. RNase T2 (▵ and ▴) cleaves 3′ of A residues. RNase V1 cleaves helical regions (a strong hit is indicated by a large arrow, a weaker hit is marked by a small arrow). The nucleotide numbers are indicated on the RNA molecule. The sequences from nt 1 to 16 and from nt 89 to 95 are derived from the cloning vector pGEM3 (Promega). Nucleotides 17–30 and 71–88 are derived from the fixed 5′-end and 3′-end sequences, respectively. The variable region extends from nt 31 to 70.