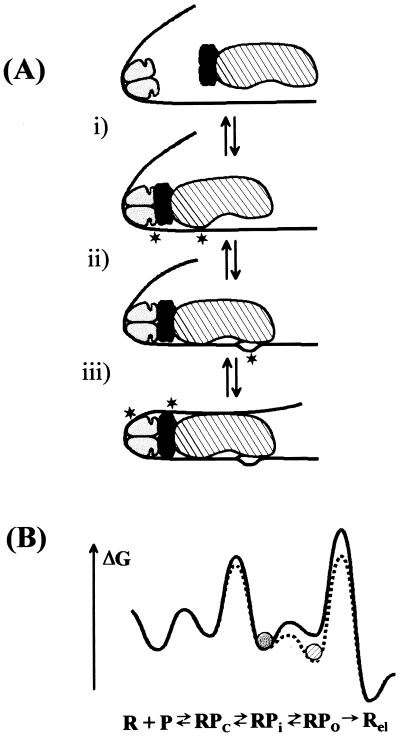
Figure 5.
(a) Model of transcriptional activation by CRP at the malT promoter. The CRP dimer is symbolized as a two-domain ellipsoid. The DNA (solid line) is bent around the prebound CRP. Newly appearing signals visible in UV laser footprinting experiments are indicated with an asterisk. The reaction is divided into three steps. (i) RNA polymerase (large ellipsoid) makes a direct contact with the prebound CRP via the carboxyl-terminal domain of the α-subunits (represented by the darker ellipsoids). Signals appear at −32 and at −60. (ii) RNA polymerase opens the DNA between the −10 hexamer and the start site of transcription. (iii) Contacts between the upstream DNA and the back of RNA polymerase stabilize the open region of DNA. The symmetry properties of the α-subunits are arbitrary; the interaction of both α-subunits with the DNA may be restricted to the stretch just upstream of the promoter. (b) Change in free energy during transcription initiation and activation. The solid line represents the malT promoter and the stippled line represents the same promoter in the presence of CRP. The diagram illustrates the redistribution of the most populated state (represented by the circles) in the presence of CRP. The rate-limiting step (highest peak) remains promoter escape under both conditions, and open complex formation is slightly accelerated in the presence of CRP. The diagram is schematic to illustrate how CRP indirectly accelerates promoter escape, and it is not meant to accurately reflect absolute free energies.