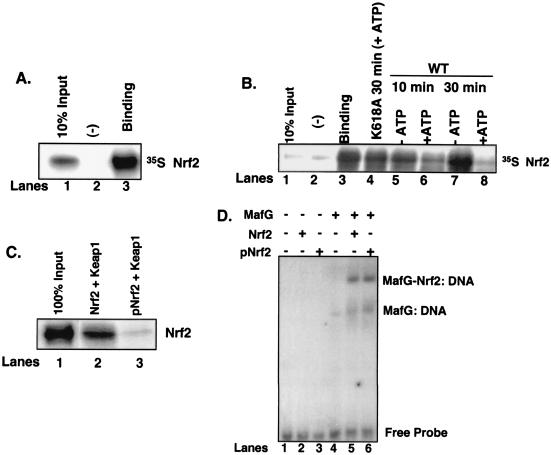
FIG. 6.
Phosphorylation-dependent dissociation of Nrf2 from Keap1. (A) Nrf2 in vitro transcribed and translated in the presence of [35S]methionine was mixed with recombinant Keap1 (lane 3) or eIF2α (lane 2). Complexes were affinity purified by nickel chromatography, resolved by SDS-PAGE, and visualized by autoradiography. Lane 1 is 10% Nrf2 input. (B) Nrf2/Keap1 complexes prepared as described for panel A and mixed with the catalytic domain of PERK in the absence (lanes 5 and 7) or presence (lanes 6 and 8) of ATP (500 μM) or a kinase-dead PERK in the presence of ATP (lane 4) for the indicated intervals. Lane 1 is 10% Nrf2 input, and lane 3 is Nrf2-Keap1 binding. Lane 2 is binding to the negative control, eIF2α. Upon completion of the kinase reactions, the complexes were washed, resolved by SDS-PAGE, and visualized by autoradiography. (C) Nrf2 was mixed with the catalytic domain of PERK in the absence (lane 2) or presence (lane 3) of ATP and then mixed with recombinant Keap1 as described above. After binding, the complexes were washed extensively and proteins were resolved by SDS-PAGE, transferred to a nitrocellulose membrane, and probed with an antibody specific for Nrf2. Lane 1 shows 100% Nrf2 input. (D) Nrf2 (lanes 2 and 5) or PERK-phosphorylated Nrf2 (lanes 3 and 6) was mixed with in vitro-transcribed and -translated MafG (lanes 5 and 6) along with a radiolabeled oligonucleotide containing a consensus ARE in an EMSA reaction. Also shown are EMSA reactions with MafG alone (lane 4) or no protein (lane 1) mixed with the radiolabeled oligonucleotide.