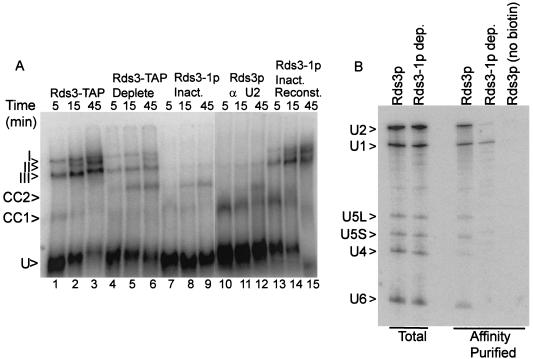
FIG. 3.
Rds3p is required for the commitment complex to prespliceosome transition. (A) Native gel electrophoresis of splicing complexes assembled on RPS17A pre-mRNA. Rds3-TAP splicing extract was assayed without additional manipulation (lanes 1 to 3) and after preabsorption with calmodulin agarose and protein A agarose affinity resins (lanes 4 to 6). Extracts were also prepared from glucose-grown, temperature-inactivated GAL1::rds3-1 yeast (lanes 7 to 9 and 13 to 15) and from wild-type yeast (lanes 10 to 12). The wild-type extract was preincubated with RNase H and an anti-U2 oligonucleotide prior to assay in order to block assembly at the commitment complex stage. In lanes 13 to 15, the extract was preincubated with pre-mRNA for 5 min prior to the addition of a 50-fold excess of unlabeled substrate and the complementing Rds3-TAP complex. The positions of the unassembled pre-mRNA (U), commitment complex bands (CC1 and CC2), prespliceosome (III), snRNP complete splicing complex (I), and catalytically active spliceosome (II) are indicated at the left. (B) Northern analysis of splicing complex snRNAs. Shown is a membrane transfer of RNA present in wild-type extract (Rds3p) and extract prepared from the glucose-grown, temperature-inactivated GAL1::rds3-1 culture (Rds3-1p dep.) probed for the spliceosomal snRNAs. RNA in the unfractionated extract (Total) is compared with that released from affinity-purified splicing complexes assembled on biotin-substituted or unmodified (no biotin) pre-mRNA. The positions of the spliceosomal snRNAs are noted at the left.