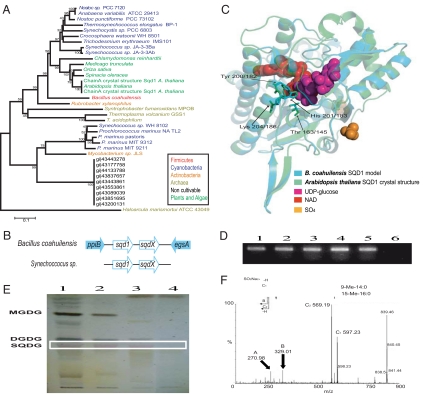
Fig. 2.
Acquisition of sulfoquinovose synthesis capabilities through HGT, an adaptation of B. coahuilensis to a phosphorus-limiting environment. (A) Neighbor-joining phylogenetic reconstruction of SQD1 (31). (B) The operon structure of the sqd1 and sqdX resembles that of the Synecchococcus genes (9). Flanking genes are ppi (peptidyl-prolyl cis-trans isomerase B) and egsA (glycerol-1-phosphate dehydrogenase) (not to scale). (C) Modeling (32) to the A. thaliana SQD (PDB ID code 1I24) protein shows conservation of the NAD and UDP-glucose-binding residues. Diagrammic representation was done by using PyMol (http://www.pymol.org). (D) Expression of the sqd1 gene. RT-PCR was carried out from RNA of B. coahuilensis grown under different phosphate concentrations (lanes 1 to 5, RT-PCR products obtained from cells cultured with 0.001, 0.005, 0.05, 0.5, and 5 mM phosphate, respectively; lane 6, control without reverse transcriptase). (E) TLC analysis reveals the presence of a probable SQDG band in B. coahuilensis, A. thaliana, Cyanobacteria sp. (lanes 1 to 3) but absent in Bacillus sp. NRRL B-14911 (lane 4). (F) Mass spectroscopic analysis confirmed the identity of the sulfoquinovoside (Fig. S3).