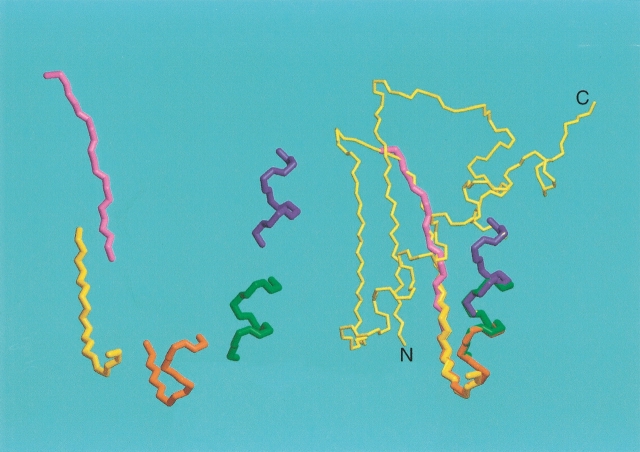
Figure 13.
Assembly of the backbone of a protein by the molecular fragment replacement (MFR) approach, using a seven-residue fragment length. For each set of seven contiguous residues, the entire PDB is searched for fragments that are compatible with the experimental dipolar couplings and chemical shifts. The 20 best-fitting fragments are minimized in a short simulated annealing protocol, and if convergence is obtained, the cluster of lowest energy structures is averaged and subsequently minimized. If only a single liquid crystalline medium is used, the orientation of each fragment relative to the alignment tensor frame is fourfold degenerate and best fitting to the preceding, partially overlapping fragment is needed to resolve this ambiguity. If dipolar couplings have been measured in two or more media, the absolute orientation of the fragment is known. Assembly is illustrated for the backbone of the RecA-inactivating protein DinI (Voloshin et al. 2001).