Figure 3.
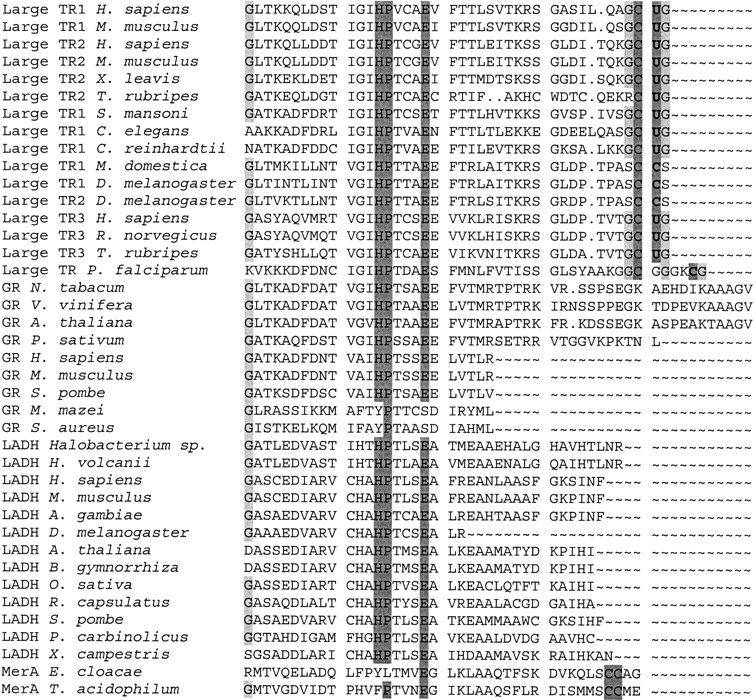
Alignment of carboxy-terminal sequences in pyridine nucleotide disulfide oxidoreductase class I family. Enzymes were aligned with BLASTP, but only carboxy-terminal regions are shown in the figure. Accession numbers for sequences are given in Figure 2B ▶. Conserved histidine, proline, and glutamate residues are highlighted in dark gray. These residues participate in the catalytic mechanism common to all pyridine nucleotide disulfide oxidoreductase class I enzymes. In addition, conserved cysteines and selenocysteines are highlighted in dark gray. These residues evolved independently in large TRs and MerAs through carboxy-terminal extensions and deletions. Glycine residues that may assist in the catalytic reaction are shown in light gray. TR, thioredoxin reductase; GR, glutathione reductase; LADH, lipoamide dehydrogenase; MerA, mercuric ion reductase.
