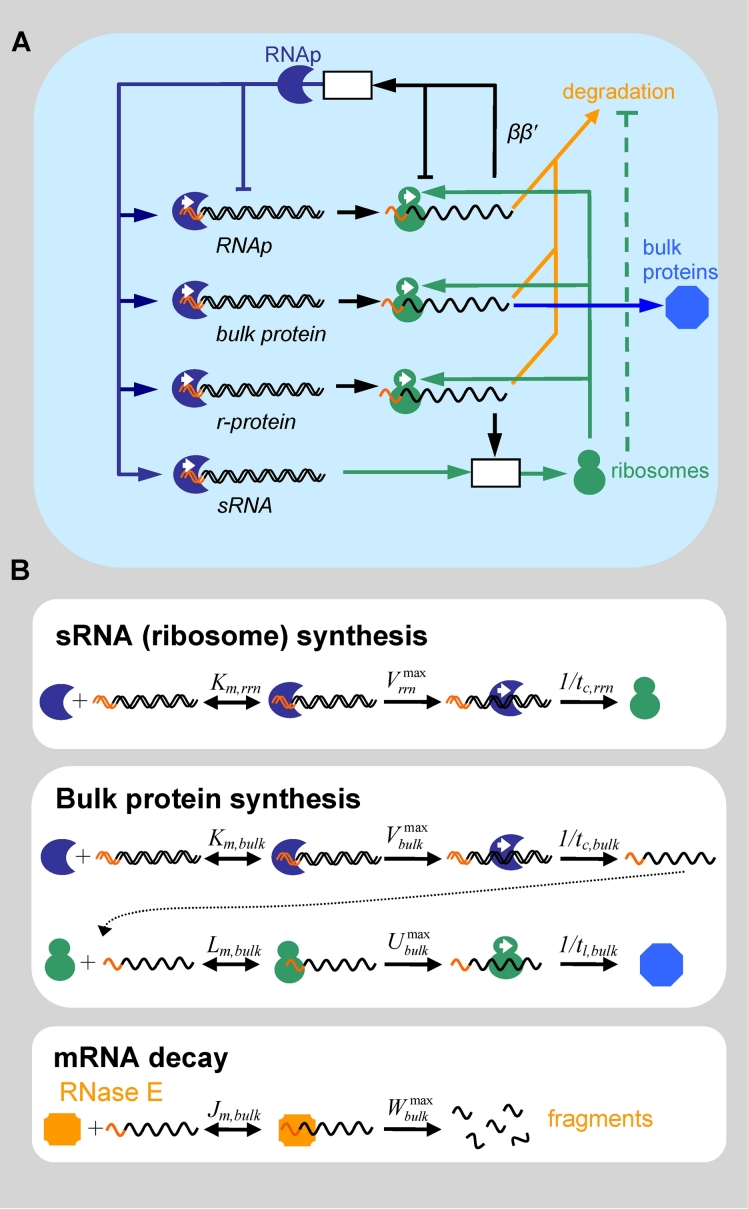
Figure 1. The coarse-grained genetic reactor (CGGR) model of E. coli.
(A) A schematic diagram of the cell as a CGGR. This figure depicts the various gene classes (double stranded DNA on the left), mRNA (single stranded RNA on the right) and their expressed products, stable RNA and the various positive (→) and negative (⊣) feedbacks between these components. The colored tips on the DNA and mRNA represent the promoter binding sites and the ribosome binding sites (RBSs) respectively. The white boxes denote the process of assembly of functional complexes from immature subunits. The light blue background represents the finite volume of the cell in which reactions take place, which is determined by the DNA replication initiation system (see text and S2.2 in Text S1). (B) The basic reactions taking place in the cell: Km
,i, 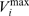 are the Michaelis-Menten (MM) parameters for transcription initiation of gene class i
[3],[28], followed by transcription at a rate of 1/tc,i. Lm
,i,
are the Michaelis-Menten (MM) parameters for transcription initiation of gene class i
[3],[28], followed by transcription at a rate of 1/tc,i. Lm
,i, 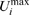 are the MM parameters of gene class i for translation initiation (i.e. binding of a 30S ribosome subunit to a RBS) [30],[31], followed by translation at a rate of 1/tl,i. Note that in this scheme, the 30S-RBS binding affinity, Lm,i, includes the 30S interaction with the secondary structure of mRNA [30]. mRNA degradation is primarily achieved via the endonuclease RNase E [35] and is assumed to follow MM like kinetics (Jm
,i,
are the MM parameters of gene class i for translation initiation (i.e. binding of a 30S ribosome subunit to a RBS) [30],[31], followed by translation at a rate of 1/tl,i. Note that in this scheme, the 30S-RBS binding affinity, Lm,i, includes the 30S interaction with the secondary structure of mRNA [30]. mRNA degradation is primarily achieved via the endonuclease RNase E [35] and is assumed to follow MM like kinetics (Jm
,i, 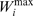 ) [36],[37]. In the scheme considered here, RNase E competes with the 30S subunits in binding to a vacant RBS [38],[39], functionally inactivating it when it binds [38]. This results in a coupling between translation and degradation [34]. Delays due to assembly are incorporated separately. The MM parameters, together with the time constants and map locations define genetic parameters for the various gene classes, which can be easily “tweaked” by base pair mutations (c.f. Table 2 and Table S1).
) [36],[37]. In the scheme considered here, RNase E competes with the 30S subunits in binding to a vacant RBS [38],[39], functionally inactivating it when it binds [38]. This results in a coupling between translation and degradation [34]. Delays due to assembly are incorporated separately. The MM parameters, together with the time constants and map locations define genetic parameters for the various gene classes, which can be easily “tweaked” by base pair mutations (c.f. Table 2 and Table S1).