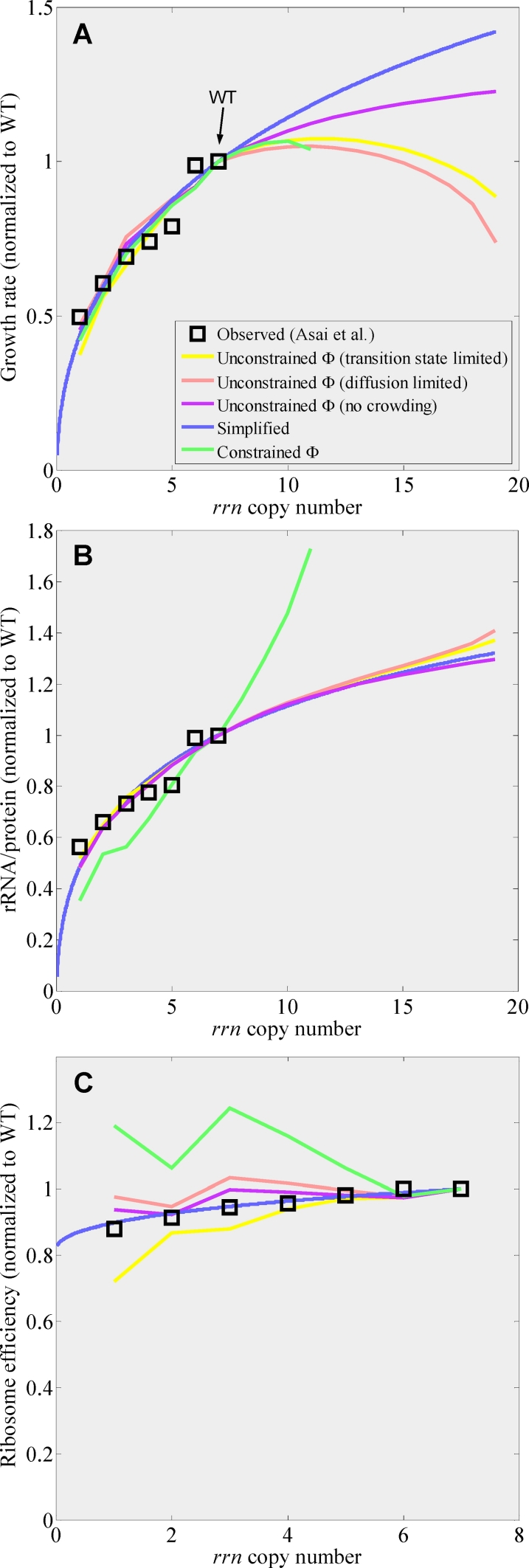
Figure 2. Comparison of rRNA operon inactivation data of Asai et al. [19] to CGGR models and predictions for higher rRNA operon copy numbers.
(A) Growth rate as a function of rRNA operon copy number per chromosome. The maximum standard error of growth rate measurements was 0.07 [19]. (B) rRNA to total protein ratio, where total protein is given by total amino acids in the form of r-proteins and bulk proteins. Measurement error was not available for this data. In the case of the constrained model, solutions were not obtainable above a copy number 11. (C) Ribosome efficiency, defined as er = α·P/Nribo [3],[19] (see text), was obtained by dividing the growth rate in (A) by the ratio of rRNA to total protein in (B). All curves are normalized to WT values at 2.0 doub/h. The legend to all figures is given in (A). The kink observed for copy number 2 is due to strong expression of lacZ in this strain used for inactivation. Beyond the WT rRNA gene dosage, rRNA operons were added at the origin (also see S1.4 in Text S1). The rRNA chain elongation rate, crrn, was assumed to be fixed in these simulations.