Figure 1.
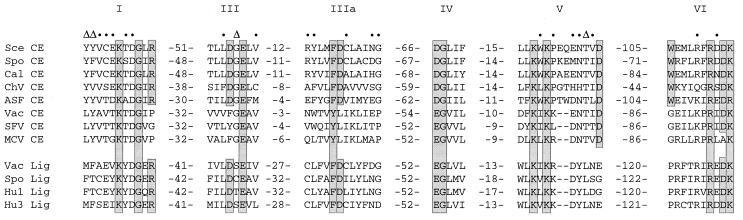
Conserved sequence elements define a superfamily of covalent nucleotidyltransferases. Six collinear sequence elements, designated motifs I, III, IIIa, IV, V, and VI, are conserved in guanylyltransferases and ATP-dependent DNA ligases as shown. The amino acid sequences are aligned for capping enzymes (CE) encoded by S. cerevisiae (Sce), Sc. pombe (Spo), C. albicans (Cal), Chlorella virus PBCV-1 (ChV), African swine fever virus (ASF), vaccinia virus (Vac), Shope fibroma virus (SFV), and molluscum contagiosum virus (MCV). Grouped below the capping enzymes are aligned sequences for the DNA ligases (Lig) of vaccinia, Sc. pombe, human ligase I (Hu1), and human ligase 3 (Hu3). The numbers of amino acid residues separating the motifs are indicated. Residues in the yeast capping enzyme Ceg1 that were found by mutational analysis to be essential for function are shown in shaded boxes. Where these residues are conserved in other family members, they are also shaded. Ceg1 residues judged to be nonessential (i.e., tolerant of alanine substitution) are denoted by dots. Ceg1 positions at which alanine replacement caused a temperature-sensitive growth defect are denoted by Δ.