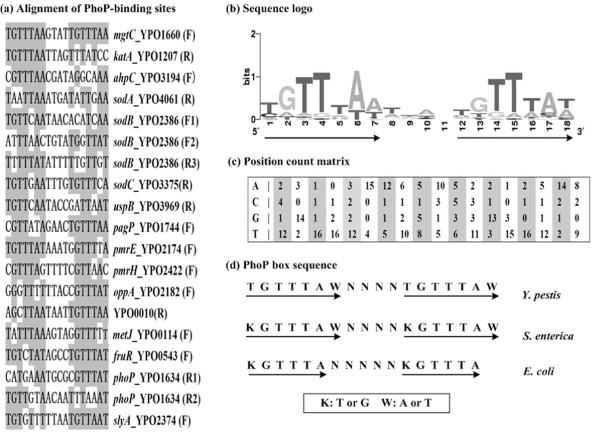
Figure 1.
Alignment of PhoP regulatory motifs in Y. pestis. (a) Homologues of the PhoP box, identified from the 19 PhoP binding sites determined by DNase I footprinting, were aligned and the conserved positions were highlighted. 'F' and 'R' in the brackets indicated the coding and noncoding stand, respectively. '1', '2' and '3' represented the numbers of PhoP-binding sites in a specific PhoP-dependent promoter (see Additional file 6 for details). (b) A sequence-Logo representation of the aligned direct repeat of the short consensus sequence (TGTTTAW). (c) Position count matrix describing the sequence logo. In the matrix, each row represents a position and each column a nucleotide. (d) An 18 bp PhoP box sequence (TGTTTAWN4TGTTTAW) of Y. pestis was generated by the sequence logo. The known corresponding ones for E. coli and S. typhimurium were shown as well.