Abstract
Comparative morphological and developmental studies, including a recent comparative study of tooth development among the Afrotherian mammals, are indicating the types of genetic mechanisms responsible for the evolution of morphological differences among major mammalian groups.
The orders of eutherian mammals are especially characterized by morphological differences in the skull and dentition, related to different requirements for processing food, and in the postcranial skeleton, which is adapted for varied modes of locomotion. The evolutionary biologist George Gaylord Simpson [1] defined major morphological discontinuities among higher taxa, specifically the orders of mammals, as the result of macroevolution or 'quantum evolution'. In many cases, these discontinuities lack fossil evidence of transitions, appearing as what Simpson termed 'breaks in the fossil record', and thus probably result from major adaptive shifts. Along with the accepted processes of microevolutionary change at the population level, Simpson also suggested that mutations with large phenotypic effects "unquestionably provide a theoretically excellent mechanism" for large changes in morphology. These discontinuities, as well as the short time periods associated with the diversification of many mammalian orders, are still presenting a challenge to paleontologists, geneticists and developmental biologists attempting to reconstruct the 'Mammal Tree of Life', a first step in understanding the geological and biological processes that are responsible for mammalian diversity [2].
For many years now, differences in gene regulation rather than dramatic differences in gene structure have been proposed as the most probable explanations for morphological and functional differences, including those between ourselves and our closest living primate relative, the chimpanzee [3]. For example, genes involved in craniofacial muscle development [4], higher brain functions [5,6], and speech and language [7] have been found to show potentially significant differences in rate of evolution or pattern of expression between chimps and humans.
Linking the Afrotheria together
The superorder Afrotheria is another challenging case of morphological discontinuity in mammalian evolution, containing animals as morphologically distinct as elephants and aardvarks. In a recent paper in BMC Biology, Asher and Lehmann [8] now provide clinching evidence for one of the few morphological and developmental traits so far identified as being common to members of this diverse group, and suggest a possible candidate gene that may repay further study.
The Afrotheria are a recently described group of African origin containing the orders Proboscidea (elephants), Sirenia (manatees and dugongs), Hyracoidea (hyraxes), Macroscelidea (elephant shrews), Tubulidentata (aardvarks), and Afrosoricida (golden moles of the family Chrysochloridae and tenrecs and otter shrews of the family Tenrecidae). Despite the obvious morphological differences distinguishing the members of this superorder (Figure 1), extensive molecular phylogenetic studies consistently support a monophyletic origin for the Afrotheria (that is, the group all descend from a single common ancestor) [9-14]. But there are few unequivocal morphological synapomorphies (shared-derived characteristics) supporting monophyly of this clade [8,15-17]. As indicated by Archibald [16], the superorder is "not predicted by fossils". This is especially the case for the Afrosoricida, whose families were once aligned with the insectivore group Lipotyphla. Novacek [17] indicates that morphologically Afrotheria is "provocative", suggesting a "radical shakeout of the placental tree".
Figure 1.
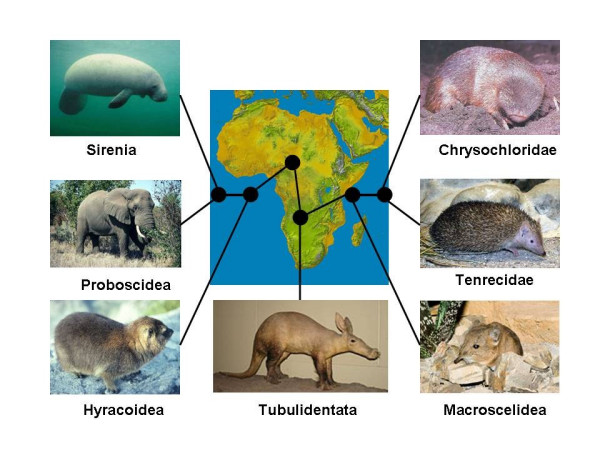
Superorder Afrotheria showing the presumed relationships among the various orders. Some of the relationships are not well confirmed. There is support for the group Paenungulata, containing Hyracoidea (hyraxes, elephants, and manatees/sea cows), Tethytheria (elephants and manatees/sea cows), Afrosoricida (families Tenrecidae (tenrecs) and Chrysochloridae (golden moles)).
Morphological investigations of Afrotheria are bearing fruit, however, as revealed by Asher and Lehmann [8], who provide evidence for the late eruption of the permanent dentition as a synapomorphy uniting the Afrotheria. It was known that tooth eruption in elephants, sea cows and hyraxes occurs only after an individual reaches its adult body size, unlike the situation in other mammals. But there was no quantitative data on dental development in the smaller Afrotherians. Asher and Lehmann [8] therefore examined the relation of jaw size to the number of permanent teeth using skulls of tenrecs and golden moles, and were able to confirm the late eruption of the permanent dentition in these animals.
Although the study of the genetics and developmental biology of the Afrotheria is in its infancy, the authors draw a comparison with a rare human developmental abnormality to identify a candidate gene that deserves further study. The human condition cleidocranial dysplasia (CCD), which disfigures the facial features, has some morphological similarities to traits held in common among Afrotheria, including late tooth eruption. Several traits similar to those associated with CCD (for example, delayed eruption of teeth, vertebral anomalies, testicondy or non-descent of male gonads, and reduction of clavicles) vary across various groups of mammals and appear to be associated with Afrotheria. Asher and Lehmann [8] used a phylogenetic context to test for covariance of these CCD-like traits, with the assumption that covariance is expected for traits controlled by the same developmental pathway. Although no significant covariance was detected, human and mouse studies do reveal that mutations in the gene Runx2, which encodes a transcription factor in the pathway controlling the development of bones and teeth, are associated with CCD [18,19], and Runx2 is a useful candidate gene for detailed comparisons across the major categories of mammals, including Afrotheria. As with many other examples, changes in gene regulation probably account for morphological similarities and differences among the Afrotheria.
Surface to air
Another, and more extensively studied, discontinuity in mammals concerns the large morphological changes that led to the evolution of flight in bats (Chiroptera) [20]. The bat forelimb represents an airfoil that results from elongation of digits, distal reduction of the radius and ulna, development of wing membranes (patagia), and modification of flight muscles and their innervation. Although morphological and molecular studies [21,22] provide a phylogenetic framework for relationships among bat families, less is known about the stages leading to the development of this airfoil and the evolution of flight. True flight undoubtedly originated early in chiropteran evolution, as the oldest fossil bat known, Onchonycteris finneyi (dated at 52.5 million years ago), has wing morphology similar to modern bats (Figure 2) [23]. By comparative studies with non-flying mammals, it is now clear that small changes in the spatiotemporal pattern of gene expression during development account for the dramatic changes represented by the chiropteran forelimb [24-27], and the genes responsible are beginning to be identified.
Figure 2.
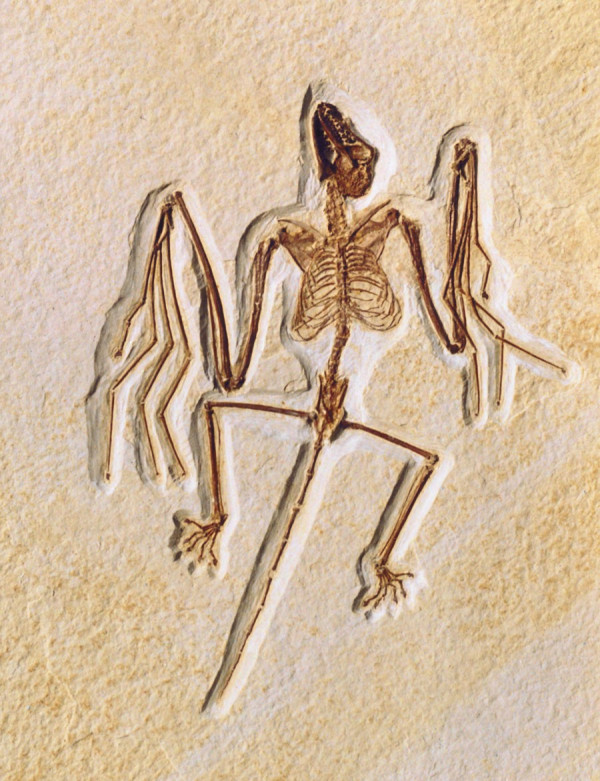
Fossil bat, Onchonycteris finneyi, collected from Green River formation in Wyoming. Photograph courtesy Nancy B Simmons.
The continued elongation of digits in bat embryos compared with mouse embryos seems to be associated with the regulation of cartilage growth [24]. One candidate gene involved in this morphological change is Bmp2 (bone morphogenic protein 2), which encodes a secreted signaling protein associated with the regulation of chondrogenesis. Expression of this gene is upregulated in bat development compared with that of the mouse [25]. Another candidate gene is Prx1 (paired-box), which encodes a transcription factor associated with growth of limb bones. A transgenic mouse with a bat Prx1 enhancer showed an increase in limb length apparently resulting from the upregulation of the endogenous mouse Prx1 gene in cartilage [26].
Separation of the digits in vertebrates involves programmed cell death of the interdigital mesenchyme. While this occurs in the bat hindlimb, it is inhibited in the forelimb, resulting in the development of the patagium. This inhibition is due to differential inhibition of the Bmp signaling pathway in the embryonic forelimb, which is also characterized by high levels of expression of the signaling protein fibroblast growth factor 8 [27]. Although the processes responsible for the evolution of powered flight in mammals are not yet known in detail, these comparative studies indicate that small changes in the timing and extent of expression in key genes can have large developmental effects [25]. Perhaps unraveling the developmental processes will provide a clearer picture of the transition from non-volant locomotion to powered flight.
Linking genotype and phenotype
A range of comparative studies, involving population genetics, genomics, proteomics, and gene-expression profiling, are now both unraveling the regulatory processes and identifying candidate genes responsible for morphological discontinuities in mammals and other organisms. Rather than simple mutations within structural genes, many of the mechanisms underlying change represent more subtle and complex changes involving gene regulation. Complex anatomical differences such as those defining the higher categories of mammals, as well as differences between more closely related species, are likely to be the result of interacting pathways that regulate gene expression during development. Changes in gene regulation seem important for a host of phenotypic differences in mammals and other organisms [28,29]. In addition, phenotypic change could result from changes such as expansion and contraction of gene families or alternative splicing of RNA transcripts. Understanding how changes in gene regulation can alter the phenotype will be considerably more challenging than investigating structural gene changes [30], and it will require a clear methodology for the identification of candidate genes as well as the dissection of pathways and networks responsible for the development of complex traits.
Whole-genome comparisons and in vivo developmental studies provide two experimental means of addressing these problems. For mammals, this means that future progress will still largely rely on well-understood model organisms such as the mouse, and on what we can learn from human pathologies [31]. The genetic hypotheses proposed for the Afrotheria and other mammals are only the beginning; in the future, an increased understanding of how regulatory changes alter phenotype should help to determine whether Simpson's hypothesis of morphological discontinuity holds up.
Acknowledgments
Acknowledgements
I thank Nancy B Simmons of the American Museum of Natural History for permission to use the photograph of Onychonycteris finneyi. Photographs of the aardvark, elephant shrew (photographer Olaf Leillinger), manatee (US Geological Survey), tenrec (photographer Wilfried Berns: CC-BY-SA-2.0-DE), and golden mole in Figure 1 were obtained from Wikipedia http://commons.wikimedia.org/wiki/Main_Page. The photograph of Africa is from NASA. Photographs of the hyrax and elephant were provided by the author.
References
- Simpson GG. Tempo and Mode in Evolution. New York: Hafner Publishing Company; 1965. [Google Scholar]
- The Mammal Tree of Life Project http://mammaltree.informatics.sunysb.edu
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- Stedman HH, Kozyak BW, Nelson A, Thesier DM, Su LT, Low DW, Bridges CR, Shrager JB, Minugh-Purvis N, Mitchell MA. Myosin gene mutation correlates with anatomical changes in the human lineage. Nature. 2004;428:415–418. doi: 10.1038/nature02358. [DOI] [PubMed] [Google Scholar]
- Rockman MV, Hahn MW, Soranzo N, Zimprich F, Goldstein DB, Wray GA. Ancient and recent positive selection transformed opioid cis-regulation in humans. PLoS Biol. 2005;3:2208–2219. doi: 10.1371/journal.pbio.0030387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, Lambert N, Lambot M-A, Coppens S, Pedersen JS, Katzman S, King B, Onodera C, Siepel A, Kern AD, Dehay C, Igel H, Ares M, Vandereghan P, Haussler D. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;44:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- Enard W, Przeworski M, Fisher SE, Lai CSL, Wiebe V, Kitano T, Monaco AP, Pääbo S. Molecular evolution of FOXP2, a gene involved in speech and language. Nature. 2002;418:869–872. doi: 10.1038/nature01025. [DOI] [PubMed] [Google Scholar]
- Asher RJ, Lehmann T. Dental eruption in afrotherian mammals. BMC Biol. 2008;6:14. doi: 10.1186/1741-7007-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer MS, Cleven GC, Madsen O, de Jong WW, Waddell VG, Amrine HM, Stanhope MJ. Endemic African mammals shake the phylogenetic tree. Nature. 1997;388:61–64. doi: 10.1038/40386. [DOI] [PubMed] [Google Scholar]
- Stanhope MJ, Madsen O, Waddell VG, Cleven GC, de Jong WW, Springer MS. Highly congruent molecular support for a diverse superordinal clade of endemic African mammals. Mol Phylogenet Evol. 1998;9:501–508. doi: 10.1006/mpev.1998.0517. [DOI] [PubMed] [Google Scholar]
- Stanhope MJ, Waddell VG, Madsen O, de Jong WW, Hedges SB, Cleven GC, Kao D, Springer MS. Molecular evidence for multiple origins of Insectivora and for a new order of endemic African insectivore mammals. Proc Natl Acad Sci USA. 1998;95:9967–9972. doi: 10.1073/pnas.95.17.9967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy WJ, Eizirik E, O'Brien SJ, Madsen O, Scally M, Douady CJ, Teeling E, Ryder OA, Stanhope MJ, de Jong WW, Springer MS. Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science. 2001;294:2348–2351. doi: 10.1126/science.1067179. [DOI] [PubMed] [Google Scholar]
- Wildman DE, Uddin M, Opazo JC, Liu G, Lefort V, Guindon S, Gascuel O, Grossman LI, Romero R, Goodman M. Genomics, biogeography, and the diversification of placental mammals. Proc Natl Acad Sci USA. 2007;104:14395–14400. doi: 10.1073/pnas.0704342104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjer KM, Honeycutt RL. Site specific rates of mitochondrial genomes and the phylogeny of Eutheria. BMC Evol Biol. 2007;7:8. doi: 10.1186/1471-2148-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher RJ, Novacek MJ, Geisler JH. Relationships of endemic African mammals and their fossil relatives based on morphological and molecular evidence. J Mammalian Evol. 2003;10:131–194. doi: 10.1023/A:1025504124129. [DOI] [Google Scholar]
- Archibald JD. Timing and biogeography of the eutherian radiation: fossils and molecules compared. Mol Phylogenet Evol. 28:350–359. doi: 10.1016/S1055-7903(03)00034-4. [DOI] [PubMed] [Google Scholar]
- Novacek MJ. Mammalian phylogeny: genes and supertrees. Curr Biol. 2001;11:573–575. doi: 10.1016/S0960-9822(01)00347-5. [DOI] [PubMed] [Google Scholar]
- Otto F, Kanegane H, Mundlos S. Mutations in the RUNX2 gene in patients with cleidocranial dysplasia. Hum Mutat. 2002;19:209–216. doi: 10.1002/humu.10043. [DOI] [PubMed] [Google Scholar]
- Aberg T, Wang X-P, Kim J-H, Yamashiro T, Bei M, Rice R, Ryoo H-M, Thesleff I. Runx2 mediates FGF signaling from epithelium to mesenchyme during tooth morphogenesis. Dev Biol. 2004;270:76–93. doi: 10.1016/j.ydbio.2004.02.012. [DOI] [PubMed] [Google Scholar]
- Simmons NB. The case for chiropteran monophyly. Am Mus Novit. 1994;3103:1–54. [Google Scholar]
- Simmons NB. Bat phylogeny: an evolutionary context for comparative studies. In: Adams R, Pederson S, editor. Ontogeny, Functional Ecology, and Evolution of Bats. Cambridge, UK: Cambridge University Press; 2000. pp. 9–58. [Google Scholar]
- Teeling EC, Springer MS, Madsen O, Bates P, O'Brien SJ, Murphy WJ. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science. 2005;307:580–584. doi: 10.1126/science.1105113. [DOI] [PubMed] [Google Scholar]
- Simmons NB, Seymour KL, Harbersetzer J, Gunnell GF. Primitive early Eocene bat from Wyoming and the evolution of flight and echolocation. Nature. 2008;451:818–822. doi: 10.1038/nature06549. [DOI] [PubMed] [Google Scholar]
- Sears KE, Behringer RR, Rasweiler JJ, Niswander LA. Development of bat flight: morphologic and molecular evolution of bat wing digits. Proc Natl Acad Sci USA. 2006;103:6581–6586. doi: 10.1073/pnas.0509716103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears KE. Molecular determinants of bat wing development. Cells Tissues Organs. 2008;187:6–12. doi: 10.1159/000109959. [DOI] [PubMed] [Google Scholar]
- Cretekos CY, Wang Y, Green ED, NISC Comparative Sequencing Program. Martin JF, Rasweiler JJ, Behringer RR. Regulatory divergence modifies limb length between mammals. Genes Dev. 2008;22:141–151. doi: 10.1101/gad.1620408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherbee SD, Behringer RR, Rasweiler JJ, Niswander LA. Interdigital webbing retention in bat wings illustrates genetic changes underlying amniote limb diversification. Proc Natl Acad Sci USA. 2006;103:15103–15107. doi: 10.1073/pnas.0604934103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabot A, Shrit RA, Blekhman R, Gilad Y. Using reporter gene assays to identify cis regulatory differences between humans and chimpanzees. Genetics. 2007;176:2069–2076. doi: 10.1534/genetics.107.073429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG. Evolutionary changes in cis and trans gene regulation. Nature. 2004;430:85–88. doi: 10.1038/nature02698. [DOI] [PubMed] [Google Scholar]
- Wray GA, Hahn MW, Abouheif E, Balhoff JP, Pizer M, Rockman MV, Romano LA. The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol. 2003;20:1377–1419. doi: 10.1093/molbev/msg140. [DOI] [PubMed] [Google Scholar]
- Carroll SB. Genetics and the making of Homo sapiens. Nature. 2003;422:849–857. doi: 10.1038/nature01495. [DOI] [PubMed] [Google Scholar]


