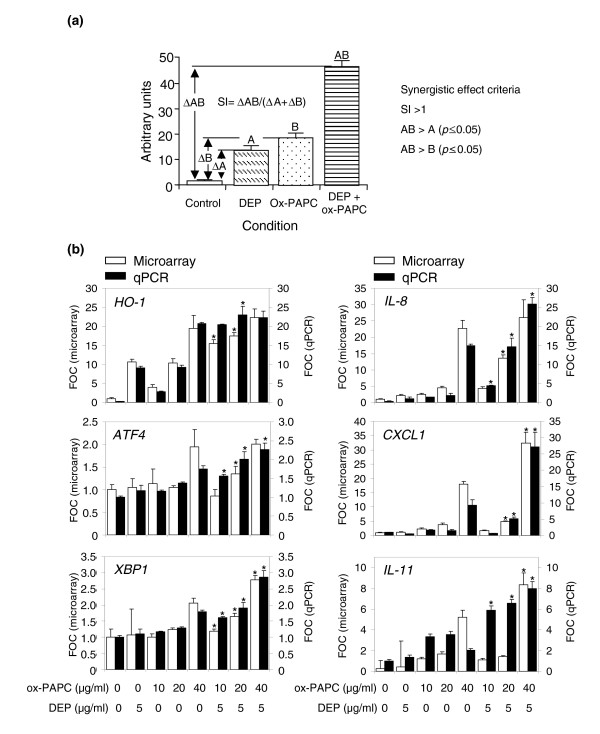
Figure 4.
DEP and ox-PAPC co-regulate genes in a synergistic/additive fashion. (a) Synergistic index (SI). Synergy was defined as the presence of a co-regulatory effect by both DEP and ox-PAPC that was greater than the effects induced by either compound alone and greater than the sum of those individual effects. The following criteria for a synergistic effect were as follows: SI (ΔAB/(ΔA+ΔB) > 1; AB (mean) > A (mean), p ≤ 0.05; AB (mean) > B (mean), p ≤ 0.05, where ΔA is the difference in mean expression level between the DEP and the control samples, ΔB is the difference in mean expression level between the ox-PAPC and the control samples, and ΔAB is the difference in mean expression level between the DEP + ox-PAPC and the control samples. (b) mRNA expression levels of representative genes. Each graph displays the relative mRNA expression levels normalized by β2-microglobulin mRNA levels and expressed as fold control (FOC) for microarray (white bars, left-hand y-axis) and qPCR (black bars, right-hand y-axis) assessment of representative genes (HO-1, IL-8, ATF4, CXCL1, XBP1, IL-11). For ease of comparison, the qPCR scale was divided by factors of 3.5 (HO-1) and 3 (IL-8), respectively. In similar fashion, the microarray scale was divided by a factor of 4 (IL-11) to make the comparison easier. The asterisk indicates combinations of DEP + ox-PAPC that exhibited synergistic effects. The high consistence of microarray and qPCR analysis, conducted on triplicate samples from independent experiments, implies both technical and biological validation. Statistical analysis was performed by one-way ANOVA, Fisher PLSD.