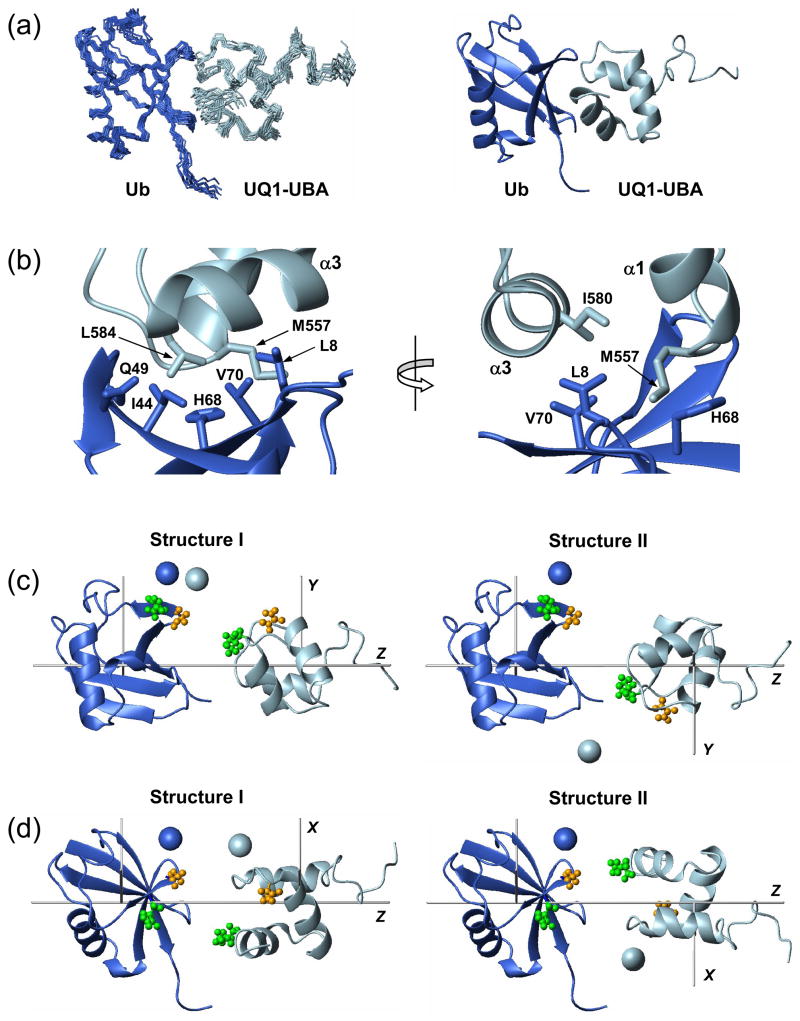
Figure 4.
The structure of the monoUb:UQ1-UBA complex. (a) The final ensemble of 10 lowest-energy NMR structures and a cartoon representation of the representative structure of the complex. (b) A close-up view of some intermolecular contacts at the Ub:UQ1-UBA binding interface. (c) Two possible relative orientations of Ub and UQ1-UBA agree with the RDC data. These conformations differ by a 180° rotation of one of the domains (in this case, UQ1-UBA) about the Z-axis of the alignment tensor; the grey rods indicate the principal axes of the tensor. Spheres represent the positions of the spin labels “reported” by the individual domains, determined from C48-spin labeling data (Fig. 3e). These structures were obtained from those in panel (a) by moving the two proteins away from each other along the Z-axis and rotating the complex around this axis by ~30°. (d) Same structures as in (c) (rotated around the Z axis) with the spin labels reconstructed from C12-SL experiments (Fig. 3f). The coloring of the SL-representing spheres in (c) and (d) is the same as in Fig. 3, i.e. the SL positions derived from measurements on SL-Ub and UQ1-UBA are colored blue and light blue, respectively. The pairs of residues that give intermolecular NOEs are shown in ball-and-stick in panels (c, d) and colored green (Q49 - L584) and orange (G47 - N561). Throughout this figure, Ub is colored blue and UQ1-UBA is light blue.