Figure 2.
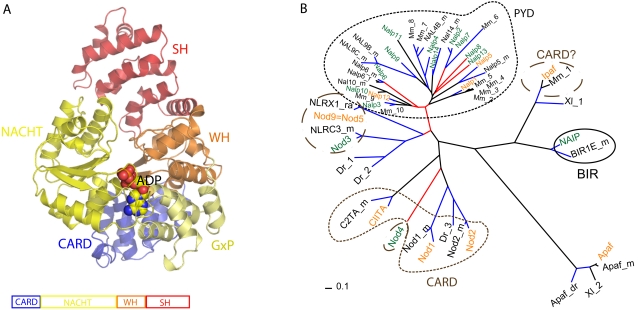
A Apaf-1 structure and domain organization. Apaf-1ΔWD40 (PDB id: 1z6t) is shown in ribbon representation color-coded according to domain boundaries (CARD aa 1–101, NACHT-GxP aa108–365, WH aa366–450, and SH aa451–586. ADP molecule bound to the active site is shown in sphere representation. B Evolutionary tree of human NLR NACHT-WH domain generated for NLR proteins and orthologues. Labels correspond to the accession numbers (Uniprot) Mm_1: Q3TAU8, Mm_2: Q2LKU8, Mm_3: Q2LKU9, Mm_4: Q2LKV8, Mm_5: A1Z198; NALP5_m: NALP5_MOUSE, Mm_6: Q4PLSO, Mm_7: Q66JP4; Mm_8: NAL4C_MOUSE, Mm_9:Q08EE9, Mm_10: Q8R4B8. Dr_1: A5PEZ1; Dr_2: A3KQD4, Dr_3: Q1AMZ9, Xl_1: Q28DS5, Xl_2: Q6GNU6. Black lines indicate a probability of more than 50%, blue lines indicate a 95% confidence in the grouping, and red lines indicate a confidence below the 40%. Green fonts indicate human proteins, black fonts alternative organisms. Mm and _m is Mus musculus (mouse), Dr and _Dr is Danio rerio (fish), Xl and _Xl is Xenopus leavis (frog). Orange fonts indicate an “S” in the Walker A motif in human sequences. Circles and modified ovals over the clades indicate the type of domain present at the N-terminal region of the NACHT domain. PYD is Pyrin domain, CARD is Card domain, Card? indicates CARD-like and BIR are BIR repeats. Apaf-1 is used to root the tree.