Abstract
Little is known about the range of hosts in which broad-host-range (BHR) plasmids can persist in the absence of selection for plasmid-encoded traits, and whether this “long-term host range” can evolve over time. Previously, the BHR multidrug resistance plasmid pB10 was shown to be highly unstable in Stenotrophomonas maltophilia P21 and Pseudomonas putida H2. To investigate whether this plasmid can adapt to such unfavorable hosts, we performed evolution experiments wherein pB10 was maintained in strain P21, strain H2, and alternatingly in P21 and H2. Plasmids that evolved in P21 and in both hosts showed increased stability and decreased cost in ancestral host P21. However, the latter group showed higher variability in stability patterns, suggesting that regular switching between distinct hosts hampered adaptive plasmid evolution. The plasmids evolved in P21 were also equally or more stable in other hosts compared to pB10, which suggested true host-range expansion. The complete genome sequences of four evolved plasmids with improved stability showed only one or two genetic changes. The stability of plasmids evolved in H2 improved only in their coevolved hosts, not in the ancestral host. Thus a BHR plasmid can adapt to an unfavorable host and thereby expand its long-term host range.
RECENTLY published analyses of prokaryotic gene and whole genome sequences have revealed that horizontal gene transfer (HGT) between closely and very distantly related Bacteria and Archaea plays a far more important role in the evolution of these organisms than had been previously recognized (Jain et al. 2002; Koonin 2003; Lawrence and Hendrickson 2003; Gogarten and Townsend 2005; Smets and Barkay 2005; Doolittle and Bapteste 2007). Moreover, many studies have provided evidence that different gene transfer mechanisms contribute to the extensive gene flux among bacteria in microbial communities (Dröge et al. 1999; van Elsas and Bailey 2002; Sørensen et al. 2005). Among these mechanisms, conjugative gene transfer mediated by so-called broad-host-range (BHR) plasmids is thought to play a very important role in gene spread among distantly related hosts (Thomas 2000). Because of their ability to transfer and replicate in quite distinct phylogenetic lineages, these extrachromosomal mobile replicons can shuffle drug resistance and many other genes among a wide range of hosts (Mazodier and Davies 1991). In spite of their importance in bacterial adaptation, such as in the rapid spread of multidrug resistance (McGowan 2006; Paterson 2006), we currently do not know if and how their host range expands or contracts over evolutionary time.
While it is obvious how BHR plasmids can improve the fitness of their host by providing it with “ready-made” genes that encode beneficial traits such as drug resistance, it is much less clear how well they persist in the absence of selection for plasmid-encoded genes. Although most BHR plasmids confer a low burden (fitness cost) to many of their hosts (Thomas 2004), highly costly plasmid carriage has been documented in a few strains (Dahlberg and Chao 2003; De Gelder et al. 2007; Heuer et al. 2007). When cells without such high-cost plasmids emerge in a bacterial population through imperfect plasmid segregation, they can quickly sweep through in the absence of selection, unless they get reinfected by the plasmid at a high enough rate (Stewart and Levin 1977; Bergstrom et al. 2000). Therefore, analogous to parasites, the most persistent and successful plasmids are those with the best inheritance system, the lowest fitness cost, and the highest infection rate (Sørensen et al. 2005). We have previously shown that the stability of a BHR plasmid is highly variable within the range of hosts in which it transfers and replicates (De Gelder et al. 2007). Within a time period of 100 generations, the model plasmid used in our previous and this present study was lost in ≥ 95% of the population in three hosts, while there was 0% detectable plasmid loss in 16 other hosts. Therefore, we define the plasmid's “long-term host range” as the range of hosts in which a plasmid is stably maintained for at least 100 generations without selection. We also designate hosts in which the plasmid is unstable or stable within this period as “unfavorable” or “favorable” hosts, respectively. It is presently not known whether BHR plasmids could evolve to adapt to some of these unfavorable hosts by improving their stability.
When plasmid–host adaptation occurs, it could represent either a host shift, whereby plasmid adaptation to one particular host negatively affects its stability in other hosts, or a true host-range expansion, when there is no trade-off between improved stability in a new host and stability in previously favorable hosts. The phenomenon of shifts in bacterial hosts has been observed for phage (Crill et al. 2000; Duffy et al. 2007; Ferris et al. 2007), but so far as we know, not for plasmids. Several very valuable experimental evolution studies have demonstrated that plasmids can adapt to a bacterial host or that the host adapts to the plasmid, but none examined evolutionary changes in the plasmid's long-term host range (Bouma and Lenski 1988; Modi and Adams 1991; Modi et al. 1991; Lenski et al. 1994; Turner et al. 1998; Dahlberg and Chao 2003; Dionisio et al. 2005; Heuer et al. 2007). Given that many BHR plasmids are involved in the rapid spread of multiple antibiotic resistance determinants, there is a need to investigate if and how these plasmids can shift or further expand their long-term host range and thus persist longer in unfavorable hosts, including potential human, animal, or plant pathogens.
While it is conceivable that a plasmid will adapt to one unfavorable host, plasmids might encounter multiple distinct unfavorable hosts within short time spans through conjugative transfer in a bacterial community. As the molecular causes of instability can be different in different hosts, plasmid mutations that increase stability in one host might be neutral or even detrimental in other hosts. Due to this form of antagonistic pleiotropy one might expect that plasmid adaptation would be different when the plasmid resides in distinct hosts over evolutionary time, as compared to when it is maintained in a single genetic background. Nothing is known about the effect of such host switches on plasmid–host adaptation.
To elucidate the ability of BHR plasmids to adapt to unfavorable hosts by improving their stability and/or fitness cost in that host, we sought to answer four questions. First, can BHR plasmids adapt to unfavorable hosts? Second, is adaptive plasmid evolution different when the plasmid is regularly switched between two distinct hosts? Third, have host-adapted plasmids merely shifted or truly expanded their host range? Fourth, what is the molecular basis of plasmid–host adaptation? To answer these questions, we performed evolution experiments with the BHR plasmid pB10 (Schlüter et al. 2003) in two hosts in which pB10 is highly unstable (De Gelder et al. 2007) under three protocols: long-term propagation in Stenotrophomonas maltophilia P21 only, in Pseudomonas putida H2 only, and alternatingly between both hosts. We then tested the stability and cost of evolved plasmids in their ancestral host. The results show that a BHR plasmid can adapt to an unfavorable host and thereby undergo host-range expansion, while regularly switching between different hosts can slightly hamper plasmid adaptation. Moreover, plasmid host-range expansion was accomplished by as little as a single mutation in a 64.5-kb plasmid.
MATERIALS AND METHODS
Culture conditions:
All experiments were carried out using Difco tryptic soy broth (TSB) or tryptic soy agar (TSA) and at 30°. All liquid cultures were incubated on a rotary shaker (200 rpm). Antibiotics were used at the following concentrations: 100 mg/liter tetracycline (Tc), 100 mg/liter amoxicillin (Amx), 50 mg/liter streptomycin (Sm), 250 mg/liter rifampicin (Rif), and 250 mg/liter naladixic acid (Nal). Media are abbreviated as follows: TSA-RifTc stands for TSA medium with rifampicin and tetracycline at the concentrations listed above. Strains and cultures were archived at −80° after mixing 1 ml of liquid culture with 0.3 ml of glycerol. Dilutions and cell suspensions were made in sterile saline (8.5 g/liter NaCl).
Bacterial strains and plasmid:
The 64.5-kb plasmid pB10, isolated from a wastewater treatment plant, is a self-transmissible, BHR IncP-1β plasmid that mediates resistance against the antibiotics tetracycline, streptomycin, amoxicillin, and sulfonamide, and against mercury ions (Dröge et al. 2000; Schlüter et al. 2003). P. putida H2 and S. maltophilia P21 were recently isolated from creek sediment and activated sludge, respectively, and were found to poorly maintain pB10 in the absence of antibiotics (De Gelder et al. 2005, 2007; Heuer et al. 2007). Other strains used were P. putida UWC1 (McClure et al. 1989), P. koreensis R28 (De Gelder et al. 2005), and Escherichia coli K12 MG1655 (ATCC 47076).
Conjugative plasmid transfer:
To transfer plasmids between strains, 2 ml of overnight-grown cultures of the plasmid donor and recipient were centrifuged, the supernatants removed, and the pellets resuspended in 200 μl TSB. Twenty μl of each mating partner cell suspension were dropped on a TSA plate on top of each other for the actual conjugation and separately as negative controls. After overnight incubation the entire cell mass of each control and conjugation mixture was harvested and suspended in 300 μl saline, from which dilutions were made to streak or plate on TSA selective for transconjugants.
Evolution experiments:
Because initial fitness increases, due to adaptation of the strains to the TSB medium, may mask mutations that improve plasmid stability or cost, strains H2 and P21 were first preadapted to TSB for 100 generations. This was done because the strains were recent environmental isolates that had not been grown in TSB before and because we have previously observed a rapid increase in carrying capacity of strain H2 within this period when grown in new medium (data not shown). One colony from a freshly streaked freezer stock was inoculated into 5 ml TSB, which was incubated for ∼16 hr, and subsequently a 4.88-μl culture was transferred into 5 ml TSB (∼10 generations per day). The same culture volume was transferred daily for 10 days (representing 100 generations of growth). A purified colony of each strain was inoculated in 5 ml TSB. After incubation, these two preadapted cultures were frozen and an aliquot was transferred to 5 ml TSB-Rif and TSB-Nal to obtain spontaneous Rif- and Nal-resistant mutants. After 24–48 hr, the cultures were turbid and an aliquot was streaked onto TSA-Rif or TSA-Nal. After incubation, one colony was restreaked, which resulted in the strains P21ancRif, P21ancNal, H2ancRif, and H2ancNal. These four strains constituted the preadapted, marked ancestral hosts without plasmid, and were archived at −80°. P21ancRif and H2ancRif were used as recipients in conjugations with an overnight-grown culture of DH5α(pB10) to obtain P21ancRif(pB10) and H2ancRif(pB10), the ancestral strains used to start the evolution experiments. In addition, the Rif-resistant hosts were used to determine stability and cost of ancestral and evolved plasmids (see further); for simplicity, they are briefly named P21anc and H2anc in the text, table, and Figures 2, 3, and 4. All four plasmid-free ancestral strains were used as recipients for evolved plasmids after every cycle of the evolution experiment.
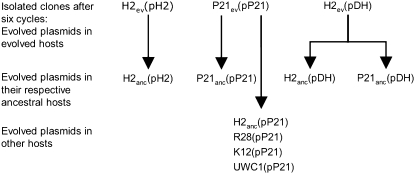
Figure 2.—
Construction of strains used for plasmid stability analysis and plasmid and strain designations are described in materials and methods and in the legend of Figure 1. All ancestral hosts used to analyze evolved plasmids were Rif resistant, but for clarity, Rif is omitted from all strain designations.
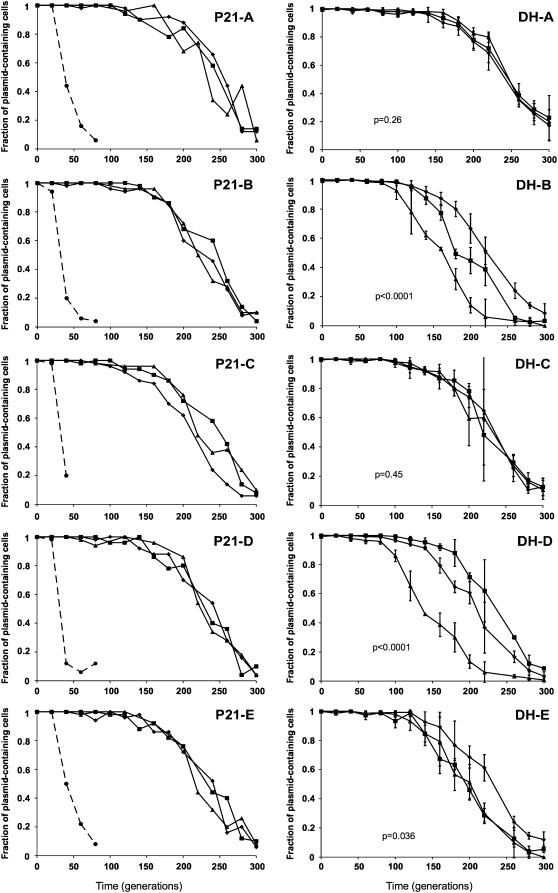
Figure 3.—
Stability of plasmids evolved in S. maltophilia P21 (pP21) and under DH protocol (pDH) in naive ancestral host P21anc. In each P21 graph (lineages A–E), the dashed line represents the stability of the ancestral plasmid in the ancestral host (one stability assay for each of the five independent ancestral cultures, A–E). In these P21 graphs, solid lines represent results from single-stability assays for three randomly chosen plasmids per lineage. In the DH graphs (lineages A–E) solid lines represent the results of triplicate-stability assays for three randomly chosen plasmids per lineage; error bars represent standard deviations. •, ▪, ♦: plasmids 1, 2, 3. Since the five DH lineages were founded from generation 70 of each of the five P21 lineages (Figure 1), ancestral plasmids for P21 and DH protocols were the same, and their stability data (dashed lines) are not repeated in the DH graphs. P-values < 0.05 ( B, D, and E) suggest that the stability dynamics were significantly different between plasmid isolates.
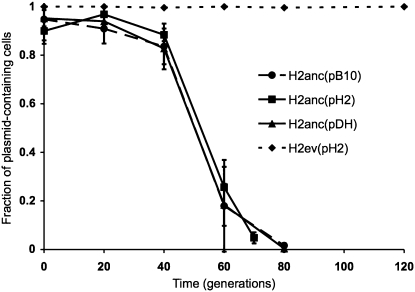
Figure 4.—
Stability of plasmids evolved in P. putida H2 (pH2) and under DH protocol (pDH), in ancestral and coevolved host H2. •, average stability of the ancestral plasmid in the ancestral host (one assay for each of the five independent ancestral cultures); ▪ and ♦, average stability of the evolved plasmids pH2 in the ancestral host (H2anc) and in the coevolved host (H2ev), respectively; ▴, average stability of the plasmids evolved in DH protocol (pDH) in the ancestral host H2anc. For all evolved plasmids, data represent averages from five evolved plasmids, one from each of the five independent lineages. Error bars represent standard deviations.
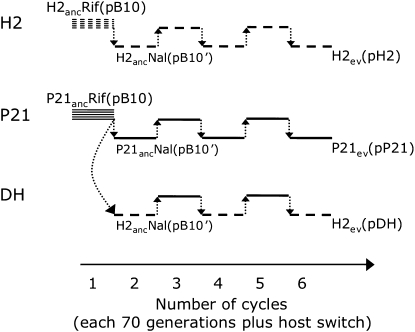
The experimental setup of the evolution experiment is depicted in Figure 1. In brief, there were three evolutionary protocols, each with five replicate lineages: Plasmid pB10 was maintained in host H2 (protocol H2), in host P21 (protocol P21), or alternatingly in both hosts (double-host protocol, DH). Approximately every 70 generations, plasmids were switched to either a genetic variant of the same ancestral host (protocols H2 and P21) or the alternate ancestral host (DH protocol) (Figure 1). We chose to switch the plasmids between strains of the same host in the H2 and P21 protocols to avoid host adaptation and promote plasmid adaptation, and to keep all parameters between the protocols identical except for the choice of host. To start the experiment, 5 separate colonies of P21ancRif(pB10) and H2ancRif(pB10) were inoculated into 5 ml TSB-Tc and incubated overnight. The 10 cultures were archived at −80°, constituting the ancestral strains of the evolution experiment (generation 0), and 4.88 μl of each were transferred to fresh 5 ml TSB-Tc medium and incubated, so that ∼10 generations were obtained per 24-hr growth cycle (1/210 dilution rate). After 70 generations of serial batch cultivation (7 days), the plasmids underwent a host switch. This was done by using the 10 cultures as donors in conjugations with overnight-grown freezer-stock cultures of the appropriate ancestral NalR strains as recipients (H2ancNal for protocols H2 and DH and P21ancNal for protocol P21, Figure 1). The resuspended cells were diluted and plated on TSA-NalTc to obtain ∼5000 small transconjugant colonies after incubation. The colonies were harvested by applying 1.5 ml of TSB-NalTc onto the plate, suspending the colonies with a spreader and transferring the suspension to a 1.5-ml microcentrifuge tube. The evolution lineages were restarted by transferring 4.88 μl of these suspensions into 5 ml TSB-NalTc. When this procedure was carried out with donor and recipient cultures separately, no visible growth was observed on the plates and the subsequent liquid media. This ensured that only transconjugants were carried through to the next cycle of the evolution experiment. These 10 cultures were grown for 7 days as described above, now in TSB-NalTc, and subsequently the plasmids were transferred by conjugation, now to the appropriate RifR ancestral host (Figure 1). The evolution experiment consisted of six such cycles of 70 generations, including five host switches (Figure 1). When assuming 20 generations of growth from a single cell to a small colony of ∼106 cells during growth of the transconjugants after each switch, the total number of generations during this experiment was estimated to be 520 (= 6 × 70 + 5 × 20). This may be a conservative estimate since this ignores possible growth of donors and transconjugants, respectively, before and after plasmid transfer during the 24-hr conjugation procedure, as well as cell death during the stationary phase in every 24-hr growth cycle.
Figure 1.—
Experimental design of the three experimental evolution protocols Plasmid evolution in single host P. putida H2 (protocol H2), in single host S. maltophilia P21 (protocol P21), and alternatingly in a double-host protocol (DH). All protocols were carried out using five independent serial batch cultures (lineages A–E), represented by the five horizontal lines in the first cycle (only one line is drawn in later cycles for clarity): – – –, H2, ___, P21. These lineages were started from five separate colonies of the ancestral host with ancestral plasmid H2ancRif(pB10) or P21ancRif(pB10), both rifampicin resistant). After 70 generations (7 days) of serial batch culture, the plasmids (designated pB10′ to indicate putative plasmid mutations) from each lineage were transferred to the nalidixic acid resistant (Nal) ancestral host (H2ancNal or P21ancNal) by conjugative transfer (dotted-line arrows). The DH protocol was initiated at this point by transferring plasmids from the five P21 lineages to the alternate host H2, thus generating lineages DH-A–DH-E. This first cycle was repeated five more times, whereby the plasmids were always transferred back into the ancestral host with the reciprocal resistance. Evolved populations at the end of cycle 6 were marked by “ev” in subscript behind the strain name.
Isolation of evolved clones and plasmids:
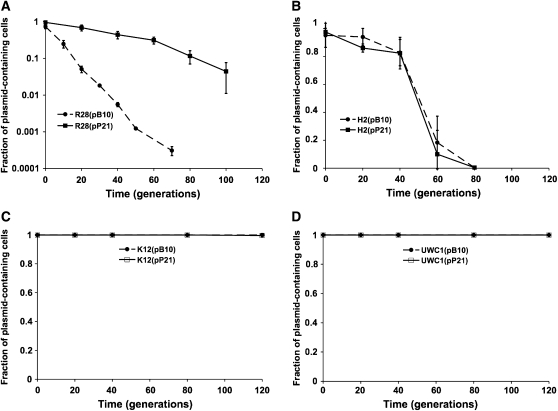
The construction of strains used for analyses is schematically depicted in Figure 2. At the end of the sixth cycle, all 15 cultures (five replicate lineages for each of the three protocols) were streaked onto TSA-NalTc, and three colonies were picked from each and inoculated into 5 ml TSB-NalTc. These 45 clones thus represented “evolved” hosts harboring evolved plasmids (Figure 2, row 1). Evolved plasmids were named on the basis of the host they evolved in: pP21 and pH 2 for plasmids that evolved, respectively, in hosts P21 and H2, while plasmids evolved in the DH protocol were named pDH. These plasmid names were followed by A–E, representing lineages A–E. Finally, the number 1 refers to plasmid 1 of three that were isolated from each lineage. Thus pP21-A1 is a plasmid from clone one in lineage A of protocol P21 and pDH-A1, a plasmid from clone one in lineage A of protocol DH. After overnight incubation, cultures founded from these evolved clones were archived and an aliquot used as donors in conjugations with the appropriate ancestral RifR hosts (Figure 2, row 2). Five plasmids evolved in P21 were also transferred to four other hosts in which they had not evolved: the ancestral P. putida H2anc, P. koreensis R28, E. coli K12 MG1655, and P. putida UWC1 (Figure 2, row 3).
Plasmid stability experiments:
Stability experiments were carried out as previously described (De Gelder et al. 2007), except for the use of TSA/TSB medium instead of LB agar/LB (Luria–Bertani). Plasmid loss was routinely assessed by replicating colonies from TSA onto TSA with and without Tc. To confirm that the loss of Tc resistance corresponded with plasmid loss and not just loss of the tet operon (De Gelder et al. 2004), some Tc-sensitive (TcS) clones were tested for sensitivity to Sm (SmS) by transferring them on TSA-Sm; true segregants should be TcSSmS. This test was done at the end point of all stability experiments depicted in Figure 3 and at each time point of the stability experiments depicted in Figure 5, C and D. The fraction of TcSSmR clones at the end of all the stability experiments in P21anc (Figure 3) ranged from 0 to 6% of the total population. This means that the fraction of plasmid-containing cells depicted in these figures, on the basis of TcS/TcR testing only, is only slightly underestimated. The fractions represented in Figure 5, C and D, are based on the fraction of TcRSmR clones and thus represent plasmid-containing cells.
Figure 5.—
Stability of plasmids pP21 (evolved in host P21) compared to stability of the ancestral plasmid pB10 in four naive hosts: two unfavorable hosts (A, P. koreensis R28; B, P. putida H2) and two favorable hosts (C, E. coli K12; D, P. putida UWC1). Data for plasmid pB10 are averages from five replicate-stability assays; data for plasmids pP21 represent averages from five evolved plasmids, one per lineage (same five plasmids that were tested in host P21anc, Figure 3). Error bars represent standard deviations. In C and D, open instead of solid squares are used for pP21 plasmids to show both symbols. Note that the y-axis of A is on a logarithmic scale. In E. coli and P. putida, both ancestral and evolved plasmids were stable for at least 200 and 300 generations (data not shown).
Plasmid cost estimation:
To estimate plasmid cost values, competition experiments were carried out as previously described (De Gelder et al. 2007), except for the use of TSA/TSB medium instead of LB agar/LB. Total and plasmid-bearing cell counts were determined on TSA and TSA-Tc plates after 1 and 2 days. Parallel control experiments starting with only plasmid-bearing cells were included to verify that no detectable plasmid loss occurred within 2 days.
Cost values for the ancestral plasmid pB10 and plasmids evolved in P21 were also estimated on the basis of the plasmid stability dynamics presented in Figure 3. We previously showed that a population dynamics model that includes plasmid loss, plasmid cost, and horizontal transfer (HT), “the HT model” adequately described the stability dynamics of ancestral plasmid pB10 in strain P21(pB10) (De Gelder et al. 2007; Ponciano et al. 2007). Here we used the same model and methods to obtain the maximum-likelihood estimates (MLEs) of plasmid cost from the stability patterns of the ancestral plasmid and the five plasmids evolved in host P21 (Figure 3). Specifically, for each of the five lineages, a set of three stability series (corresponding to the three plasmids per lineage) was treated as three replicates of the same process. For the ancestral strain, we used the five replicate stability curves to obtain one average value. Parametric bootstrap (PB) confidence intervals for the HT-model cost estimates were computed as previously described (De Gelder et al. 2004; Ponciano et al. 2007). The point estimates (MLEs) and PB error bounds (LCL and UCL for lower and upper confidence interval limits) were compared to the empirical cost estimates and their error bounds (Table 1). Finally, likelihood ratio tests for the HT model were carried out, where under the null hypothesis, the observed data were binomially distributed with a mean equal to the deterministic model predictions.
TABLE 1.
Plasmid cost (expressed as % decrease in host fitness) of ancestral and evolved plasmids in ancestral host P21anc
| Lineage | Cost of pB10 [mean (LCL, UCL) (%)] | Cost of pP21 [mean (LCL, UCL) (%)] | Model-derived cost of pP21 [MLE (LCL, UCL) (%)] | Cost of pDH [mean (LCL, UCL) (%)] |
|---|---|---|---|---|
| A | 43.2 (37.2, 49.2)a | 10.3 (6.8, 13.7) | 4.8 (4.06, 6.85) | 13.6 (8.9, 18.4) |
| B | 50.7 (46.5, 55.0) | 9.1 (6.8, 11.5) | 4.9 (4.48, 7.90) | 14.1 (10.9, 17.3) |
| C | 42.0 (40.5, 43.5)a | 10.2 (6.9, 13.5) | 4.3 (3.94, 7.56) | 10.6 (4.6, 16.6) |
| D | 49.7 (41.9, 57.6) | 12.5 (7.0, 18.0) | 4.8 (4.42, 5.31) | 12.3 (6.3, 19.3)a |
| E | 45.0 (39.4, 50.6) | 10.1 (3.9, 16.2)a | 5.0 (4.27, 8.56) | 13.3 (7.9, 18.8)a |
| Average | 46.1 (44.5, 47.8) | 10.4 (9.9, 11.0) | 12.8 (12.2, 13.4) |
Plasmid cost was determined in two ways. First, cost was measured through competition experiments between the plasmid-bearing and plasmid-free ancestral P21 strain (columns 2, 3, and 5). The plasmids used correspond to plasmid 1 in all of the Figure 3 graphs. For each plasmid, the mean plasmid cost was calculated from five replicates (except for data with only four replicates). To compare the data with model-derived estimates, the lower and upper limit (LCL and UCL) of the 95% confidence interval are presented. The average cost of ancestral and evolved plasmids was calculated from the means of all lineages. Second, the cost of pP21 plasmids was also estimated on the basis of the stability data (Figure 3) and the deterministic HT model, as in Ponciano et al. (2007) (column 4). MLE, maximum-likelihood estimate; LCL and UCL, the lower and upper limits of parametric bootstrap confidence intervals. For the ancestral plasmid, the MLE of the model-derived cost of the plasmid (based on one stability pattern for each of the five ancestral plasmid replicates (A–E) was 54.6% (33.3%, 126%).
Mean plasmid cost calculated with only four replicates.
Statistical analyses:
To verify whether the stability of different plasmids within a lineage was statistically different, we used a repeated measures ANOVA to analyze the data per lineage. In the model we took as fixed effects the factors plasmid and day. The replicate was considered a random effect. The model equation is
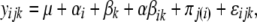 |
where αi corresponds to the plasmid effect, βk corresponds to the effect of time (day), αβik is the interaction between plasmid and day, πj(i) the random effect due to the jth replicate of plasmid i, and ɛ the residuals. Only days for which data is available for all assays were included into the analyses. The response variable yijk is the square root of the fraction of plasmid-free cells per day, per replicate, per plasmid. These analyses were implemented in statistical analysis software.
Plasmid DNA sequencing:
The complete nucleotide sequence was determined for four evolved plasmids: pP21-B1, pP21-D1, pDH-B1, and pDH-D1 (see Isolation of evolved clones and plasmids above). The evolved plasmids were first transferred to E. coli K12 MG1655 by conjugation. Cultures used for plasmid extraction were grown in LB-Tc10. Plasmid DNA was extracted using the Plasmid Mini kit (QIAGEN, Valencia, CA) according to manufacturer's instructions and using the recommendations for low-copy-number plasmids. Approximately 95% of each plasmid was sequenced by Macrogen (South Korea) using pyrosequencing technology with ∼20× coverage. To close the gaps and to examine every potential mutation as suggested from the pyrosequencing data, additional sequence determination was done in-house using a 3730 DNA analyzer (Applied Biosystems, Foster City, CA). PCR and sequencing primers were designed using Primer3. PCR was performed using AccuPrime Pfx DNA Polymerase (Invitrogen, Carlsbad, CA) according to manufacturer's instructions. Applied Biosystems Big Dye Terminator v3.1 cycle sequencing kit was used for the sequencing reactions, from which the DNA was purified using DyeEx 96-kit or 2.0 (QIAGEN). Sequence data were analyzed using NCBI Blast and ContigExpress (Vector NTI).
RESULTS
Stability of evolved plasmids in their ancestral hosts:
Our first goal was to determine if plasmids had adapted to their hosts by improving their stability, after being maintained under selection for ∼500 generations. Therefore, we tested the stability of evolved plasmids in the ancestral hosts in the absence of selection. The plasmids evolved in S. maltophilia P21, named pP21, were much more stable in the ancestral host P21anc than the ancestral plasmid pB10 (Figure 3, left, P21-A–P21-E). Thus genetic changes that affect segregative loss rate, plasmid cost, and/or conjugative transfer, must have occurred in these plasmids. Moreover, all 15 plasmids, 3 from each of the five independently evolved replicate lineages, showed strikingly similar plasmid stability dynamics. The data demonstrate that a plasmid can drastically improve its stability in an initially unfavorable host within ∼500 generations.
To examine if plasmid adaptation to unfavorable host P21 would be different when the plasmid was regularly switched between two hosts than when it is maintained in one host, we compared the stability of plasmids evolved in P21 with those evolved alternatingly in hosts P21 and H2 (named plasmids pDH). Because of the expected variation between and within lineages, triplicate stability assays were performed for each of the three plasmids per lineage. While the plasmids evolved under this DH protocol also showed increased stability in host P21anc compared to pB10, in some of the lineages the stability patterns displayed much more variation and some plasmids were less stable than those evolved in host P21 alone (Figure 3, DH graphs vs. P21 graphs). Moreover, within three lineages (B, D, and E) there was a statistically significant effect of the plasmid replicate (P < 0.05) on the stability pattern (see P-values in Figure 3), suggesting the presence of distinct plasmid mutants in the population. Overall, these results show that switching the plasmid between two unfavorable hosts can hamper plasmid adaptation in some populations.
The plasmids that evolved in host P. putida H2 (named pH2) did not show improved stability in the ancestral host H2anc compared to the ancestral plasmid pB10 (Figure 4). This shows that during 500 generations no genetic changes in the plasmid had been selected that measurably increased plasmid stability in the ancestor. Similarly, the stability of the pDH plasmids in H2anc was also not different from that of pB10. However, the clones of evolved H2 strains, H2ev(pH2) showed highly increased plasmid stability (Figure 4, horizontal dashed line). Thus within 70 generations the host adapted to the plasmid, and/or plasmids and hosts coevolved. Detailed analysis of this (co)evolutionary process was beyond the scope of this study, but could reveal new pathways of plasmid host-range shifts.
Cost of evolved plasmids:
Plasmid cost is one of the factors that determine plasmid stability patterns, because it affects the rate at which plasmid segregants sweep through the population in the absence of selection. To determine whether the increased stability of the plasmids that evolved under the P21 and DH protocols was in part due to a decrease in plasmid cost, competition experiments between plasmid-carrying and plasmid-free ancestral P21 strains were performed in antibiotic-free medium (Table 1). Since the competition was done for only 20 generations, plasmid loss was not yet significant (Figure 3) and did not confound the results. The average cost of the ancestral plasmid to the ancestral host, tested in five replicate clones, was 46%, an extremely high value. In contrast, all evolved plasmids showed a significantly lower cost to the ancestral host (P < 0.0001). Only for one of the five lineages (B), the cost of plasmids evolved in the DH protocol (pDH-B) was significantly higher (P = 0.008) than that of the plasmids from the corresponding P21-B lineage. However, when pooling all the data from each protocol together over lineages, the cost of all plasmids isolated from the DH protocol was significantly higher than the cost of all plasmids evolved in P21 (P = 0.024). Thus during long-term association of plasmid pB10 with single host P21, the plasmid reduced its cost to that host significantly, and this cost reduction was also significant but slightly less pronounced when the plasmid was regularly switched between two distinct hosts.
To validate the drastic decrease in plasmid cost after adaptation to host P21, as determined by competition assays, we also estimated the cost of the ancestral and pP21 plasmids on the basis of the stability curves shown in Figure 3 (P21-A–P21-E), using a mechanistic time-series model (De Gelder et al. 2007; Ponciano et al. 2007). Very much like the competition-derived plasmid cost values, the model-derived cost estimates were much lower for the evolved plasmids than for the ancestral plasmid. Moreover, there was no statistically significant difference between both types of cost estimates (Table 1). The likelihood ratio test results provided strong support for the deterministic HT model for three of the evolved plasmids (P21-B, -D, and -E), but not for the ancestral plasmid or P21-A and P21-C (data not shown). While rejection of this deterministic null model in three of the six data sets suggests that the predicted mean trend and/or the variance are not well explained (De Gelder et al. 2004), the results strengthen our conclusion that a plasmid can drastically improve its long-term stability by reducing its cost.
Plasmid host-range expansion:
To determine whether the plasmids that evolved in a single host, S. maltophilia P21, merely shifted or truly expanded their host range, plasmid stability was tested in four “naive” hosts, which had not previously carried plasmid pB10 (Figure 2, row 3). In two of these hosts, P. putida H2 and P. koreensis R28, the ancestral plasmid pB10 was known to be unstable, whereas in the other two, E. coli K12 MG1655 and P. putida UWC1, it is very stable (De Gelder et al. 2007; Ponciano et al. 2007). Surprisingly, the evolved plasmids were much more stable than pB10 in the unfavorable host R28 (Figure 5A). In contrast, the stability of these plasmids in strain H2 did not differ from that of pB10 (Figure 5B). This shows that the adaptive changes in the evolved plasmids that were responsible for increased stability in host P21 also improved stability in naive host R28, but not in H2. In the two favorable hosts the evolved plasmids were at least as stable as pB10 (Figure 5, C and D). On the basis of these results, these plasmids did not undergo an obvious evolutionary trade-off typical for a host-range shift, but truly expanded the range of hosts in which they can be stably maintained.
Mutations in evolved plasmids:
To determine the molecular basis of the observed improvement in plasmid cost and stability, four plasmids were completely sequenced: pP21-B1, pP21-D1, pDH-B1, and pDH-D1. These four plasmids were chosen because pDH-B1 and pDH-D1 showed the least improvement in stability compared to all other evolved plasmids (lowest stability curves in Figure 3, DH-B and DH-D). Thus they differed most from their counterpart plasmids evolved in P21 alone, pP21-B and pP21-D. Because there was no notable difference in stability and cost among these pP21 plasmids, one was randomly chosen from each of the two lineages (and named pP21-B1 and pP21-D1, respectively). Interestingly, all four plasmids shared the same genetic change: a point mutation in trbC that resulted in an amino acid change from valine to alanine in the TrbC protein at amino acid 95, which is located in the transmembrane region. The trbC gene codes for a putative prepilin, which is involved in mating-pair formation. This mutation was the only one in the entire 64.5-kb genomes of the two plasmids evolved in P21 and thus must be the cause of the observed increase in plasmid stability and cost. The mutated plasmids can still transfer from P21 to various hosts (strains P21, H2, R28, K12, and UWC1), but a difference in transfer frequencies (transconjugant/recipient fractions after overnight mating on LB agar) was observed as follows. When K12 was the recipient and P21Rif the donor, the frequency for pP21-D1 was ∼10-fold lower than for pB10. However, when P21Nal was the recipient strain, in matings either with P21Rif or with E. coli K12 as donor, evolved plasmid pP21-D1 transferred at an ∼1000-fold higher frequency than pB10 (data not shown). This increased transfer frequency may help explain how this mutant plasmid rapidly swept through the population, since it would outcompete pB10 in the conjugative transfer during every host switch.
The sequence data for the two pDH plasmids showed a second genetic change, a duplication of the orfE-like integron gene cassette, resulting in two and three copies of this cassette in pDH-B1 and pDH-D1, respectively. To confirm this interesting gene cassette amplification, PCR was performed using primers 5′CS and 3′CS that specifically anneal to the 5′- and 3′-conserved sequences of the class 1 integrons (Lévesque et al. 1994; Szczepanowski et al. 2004). The PCR products of pB10, pDH-B1, and pDH-D1 were ∼1.3 kb, 1.8 kb, and 2.2 kb, respectively, which corresponded to the sizes expected for one, two, or three copies of the orfE-like cassette. Since the genes in the integron are under the control of a general promoter, we examined if this gene duplication affected expression of oxa-2, an integron gene that is located downstream of orfE and encodes resistance to β-lactam antibiotics. Amoxicillin resistance of K12(pDH-B1) and K12(pDH-D1) was tested in LB-Amx, and both strains had lost the resistance. It is currently not known if and how this genetic change in the integron contributed to improved plasmid cost and stability in hosts H2 and/or P21, but loss of expression of the oxa-2 gene is one possible explanation. The lower stability of these two pDH plasmids in P21anc, compared to plasmids P21-B1 and pP21-D1, suggests that the mutation was detrimental to stability in host P21.
In conclusion, the clear example of parallel evolution of the trbC mutation, which was the only mutation in plasmid evolved in host P21, strongly suggests that this mutation was responsible for the improved plasmid cost and stability and was under very strong selection in both the P21 and DH evolution protocols. The molecular analysis also showed that one or two genetic changes in the genome of a BHR plasmid can expand its long-term host range.
DISCUSSION
Despite the threat of drug resistance and virulence plasmids to human, animal, and plant health, we do not understand if and how the host range of a plasmid evolves over time. In this study we examined if a BHR multidrug resistance plasmid, the IncP-1 plasmid pB10, could adapt to two unfavorable hosts through evolutionary changes in its genome. Adaptive evolution of plasmid pB10 during ∼500 generations in host P21 resulted in improved stability and decreased cost in this host. Plasmids that were maintained alternatingly in both hosts, also showed improved stability and cost in ancestral host P21anc, but rapid adaptation was hindered by regular host switching. In contrast, during that same evolutionary time, the same plasmid did not adapt to the second host, H2, but the host had adapted to the plasmid. Plasmids adapted to host P21 were also more or equally stable in other naive hosts compared to ancestral plasmid. This suggests for the first time that a BHR plasmid can even further expand its long-term host range. DNA sequence analysis of evolved plasmids indicates that one genetic change in a 64.5-kb genome caused the drastic improvement in plasmid cost and stability. This work provides new insights into the adaptability of BHR plasmids to initially unfavorable hosts, and in the possible mechanisms of evolution of their long-term host range.
There was a striking similarity in the stability dynamics and fitness-cost values of the evolved plasmids that were randomly picked from the five independently evolved lineages of S. maltophilia P21(pB10) (Figure 3 and Table 1). Although phenotypic convergence could have arisen through different adaptive walks in genotype space, DNA sequence analysis of evolved plasmids suggests parallel evolution, since a single point mutation in the trbC gene independently swept through at least two of the five P21 lineages. In a previous study we demonstrated that the high plasmid cost in host P21 was the main reason for high instability of pB10 in this host (De Gelder et al. 2007). Our results here show that a drastic improvement in stability of the evolved plasmid was accompanied by a significant decrease in the cost of the plasmid to the host. The cost, calculated on the basis of competition experiments, decreased from 46% for pB10 to 9–12% for the plasmids evolved in P21. These results were corroborated by the plasmid cost estimates on the basis of stability patterns, obtained with the horizontal transfer model developed earlier (De Gelder et al. 2007; Ponciano et al. 2007) (a decrease in cost from 54 to 5%). The single mutation in the prepilin protein (TrbC) in two independently evolved plasmids pP21-B1 and pP21-D1, strongly suggests that this amino acid change must be responsible for the improvement in cost and stability. Future studies will have to determine how a change from Val to Ala in the TrbC protein can explain such a drastic effect on plasmid cost and stability in this host. The higher frequency of transfer of evolved plasmids like P21-D1 into host P21 in filter matings compared to the poor transferability of the ancestral plasmid, may in part be responsible for the higher stability in liquid medium. However it is unlikely to explain the improved cost as determined by competition experiments in liquid medium, because IncP-1 plasmid transfer in liquids is typically too low to confound the competition experiment (Bradley et al. 1980). Moreover, the mathematical model used to estimate plasmid cost would have to be wrong in its prediction of the parameter values corresponding to the underlying causes of plasmid stability. In spite of the uncertainty about the molecular mechanism of improved plasmid cost and stability, our results suggest that a single plasmid-encoded mutation explains the drastically improved plasmid stability in independently evolved lineages of S. maltophilia P21(pB10).
We further demonstrated for the first time that regular horizontal transfer of a plasmid between distinct unfavorable hosts can have a negative effect on plasmid adaptation to a novel host. For three of the five DH lineages (DH-B, DH-D, and DH-E), three randomly chosen plasmids per population showed significant differences in stability dynamics. These results indicate the coexistence of and competition between subpopulations that harbored genetically distinct plasmids with different stability characteristics. We therefore conclude that clonal interference in three of the five lineages slowed down fixation of plasmid-encoded stability-enhancing mutations in these large populations, as previously suggested in theoretical models and experimental studies of bacteria (Gerrish and Lenski 1998; De Visser and Rozen 2006). These findings suggest that “generalist” plasmids, which frequently transfer between distinct hosts in natural communities, would not adapt as fast to any host as “specialist” plasmids that remain in one host. This is consistent with theory that specialist species, which have narrower niche breadths than generalists, are able to evolve faster than generalists (Whitlock 1996). In spite of this constraint on the evolution of generalist plasmids, BHR plasmids like those of the IncP-1 group seem to compete successfully with narrow-host-range (NHR), specialist plasmids, as they are found in many diverse hosts worldwide (Schlüter et al. 2007). However, as we recently showed (De Gelder et al. 2007), these plasmids are not necessarily stable in all hosts within their replication range. Thus they may not be very well adapted to a subset of hosts but nevertheless persist due to their high horizontal transfer (infection) rate (Bergstrom et al. 2000).
There are at least three explanations for the higher variability in stability patterns of plasmids evolved under the DH protocol compared to those evolved in single host P21. First, under that regime plasmid pB10 was maintained in host P21 for only 50% of the evolutionary time compared to when evolved in P21 only, while it spent the other 50% of the time in host H2. Therefore, mutations that were adaptive in P21 but neutral in H2 might have been occasionally lost by drift during propagation in H2 or may not have swept through the populations yet at the end of our experiment. Second, beneficial mutations selected for in P21 might have been under negative selection while in host H2 due to negative effects on plasmid stability or cost in that host (an example of antagonistic pleiotropy in heterogeneous environments). They would thus have a low chance of surviving the next severe bottleneck when switched back to P21. Since plasmids adapted to P21 showed the same stability patterns in host H2 as the ancestral plasmid (Figure 5A), it is more likely that the plasmid mutations were neutral and not deleterious for stability in host H2. Third, antagonistic pleiotropy may have played a role because mutations selected during the cycle in host H2 may have been deleterious to host P21. Our results support this last hypothesis since the sequenced plasmids pDH-B1 and pDH-D1, which have multiple copies of orfE in the integron in addition to the TrbC mutation, showed the lowest stability in P21anc (represented by the lower stability curves in Figure 3, DH-B and DH-D). Moreover, this genetic change was observed only in the two sequenced plasmids that were maintained in both hosts, and not in the two pP21 plasmids. In addition, the complete sequence of one plasmid evolved in host H2 in a previous study (Heuer et al. 2007) also contains a change in orfE, i.e., an almost complete deletion of the ORF (J. Williams and E. Top, unpublished data). More data are needed to understand the effect of the integron rearrangement, but it could represent an example of antagonistic pleiotropy with respect to plasmid stability.
Our results obtained for switching pB10 between hosts P21 and H2 are not necessarily representative of all adaptive scenarios during plasmid host switching in general. For example, if strains P21 and R28 had been chosen as plasmid hosts for this experiment, plasmid adaptation during the DH protocol might have been very similar to that in the single host protocols, as mutations in P21 also clearly improved plasmid stability in R28 (Figure 5, and see discussion below). Thus with respect to molecular plasmid–host interactions that affect plasmid stability, those two hosts may represent more similar environments for the plasmid. Overall, we can conclude that plasmid adaptation to one unfavorable host may be hampered when the plasmid is regularly switched over evolutionary time between genetically distinct unfavorable hosts compared to when it is maintained in only one host.
The plasmids that adapted to host P21 by improving their stability and cost, showed equal or improved stability in four naive hosts. This finding suggests that they truly expanded their long-term host range. The most intriguing result is the improved stability of pP21 plasmids in the naive host R28, which suggests that the trbC mutation must benefit plasmid stability in more than one host. In the absence of selective pressure, these plasmid variants with expanded host range might become more dominant in the bacterial community as they are now stable in a wider range of hosts, thus representing more successful mobile elements. We are not aware of any similar findings in the literature, but at least two studies corroborate our results. Dionisio et al. (2005) grew E. coli K12 with the NHR drug resistance plasmid R1 for 420 generations. Whereas the ancestral plasmid conferred a high fitness cost to the E. coli host, evolved plasmids conferred a fitness advantage not only to this host, but also to a naive Salmonella enterica strain. Thus, while stability was not examined in that study, the improved fitness in a naive host would probably lead to longer plasmid persistence in the absence of selection (antibiotics) compared to ancestral plasmid R1. The molecular basis of the fitness increase was not described. In addition, in vitro mutagenesis studies have shown that the replication range of the NHR P. syringae plasmid pPS10 can be expanded to more than one host by a single genetic change in the plasmid RepA protein (Fernandez-Tresguerres et al. 1995; Maestro et al. 2003). Further studies are needed to improve our insight into the tempo and mechanisms of plasmid host-range shift or expansion.
In contrast to the very clear plasmid adaptation to host P21, plasmids that evolved in host H2 did not show any detectable improvement in stability in the ancestral host. However, evolved H2 strains maintained their plasmids for at least 200 generations (Figure 4). This suggests that host evolution (or plasmid–host coevolution) has occurred in these lineages within one cycle since the last host-switching event, thus during 70 generations. Given this short time period, the underlying mechanism is most likely a single genetic change in the host chromosome. The poor stability in H2anc of plasmids evolved in this host is contradictory to our previous finding that genetic changes in pB10 resulted in drastic improvement of plasmid cost and stability in host H2 (Heuer et al. 2007) (J. Williams and E. Top, unpublished data). However, the experiment was done in a different medium and under different bottleneck and host-switching regimes. Not only was the daily dilution rate for the batch cultures in this study higher than previously (1/1000 vs. 1/256), but host switching by conjugative plasmid transfer every 70 generations (vs. every 100 previously) was done using a different procedure than before. Both changes resulted in stricter bottlenecks in the current study. Thus, our results show that the potential of a BHR plasmid to adapt to unfavorable hosts depends on the host (P21 vs. H2 in this study), as well as on the environmental conditions or population dynamics [H2 in this study vs. Heuer et al. (2007)]. Moreover, although preliminary, our study suggests that a single host mutation may allow stable maintenance of a previously unstable plasmid. This corroborates our previous finding that subtle differences in host genotypes among strains of the same species can affect plasmid stability (De Gelder et al. 2007; Sota and Top 2008). It is also in agreement with the observation that a single point mutation in DnaA of E. coli allowed replication of the NHR P. syringae plasmid pPS10 in this host (Maestro et al. 2002). Future analysis of the evolved H2 hosts will improve our currently poor understanding of the role of plasmid–host interactions in plasmid stability.
Acknowledgments
We thank L. Forney for valuable suggestions about the experimental design. We are also indebted to the two reviewers for their insightful comments, which helped us to improve this manuscript. We are grateful to S. Bassler and S. Sax for their assistance in the experiments, M. Bauer and L. Rogers, for the final touches on the experimental work, and C. Brown for assistance with sequence analysis. This project was supported by the National Institutes of Health grants no. P20 RR-16448 from the COBRE Program and no. P20 RR-016454 from the INBRE Program, both of the National Center for Research Resources.
References
- Bergstrom, C. T., M. Lipsitch and B. R. Levin, 2000. Natural selection, infectious transfer, and the existence conditions for bacterial plasmids. Genetics 155 1505–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouma, J. E., and R. E. Lenski, 1988. Evolution of a bacteria/plasmid association. Nature 335 351–352. [DOI] [PubMed] [Google Scholar]
- Bradley, D. E., D. E. Taylor and M. L. Cohen, 1980. Specification of surface mating systems among conjugative drug resistance plasmids in Escherichia coli K12. J. Bacteriol. 143 1466–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crill, W. D., H. A. Wichman and J. J. Bull, 2000. Evolutionary reversals during viral adaptation to alternating hosts. Genetics 154 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlberg, C., and L. Chao, 2003. Amelioration of the cost of conjugative plasmid carriage in Eschericha coli K12. Genetics 165 1641–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gelder, L., J. M. Ponciano, Z. Abdo, P. Joyce, L. J. Forney et al., 2004. Combining mathematical models and statistical methods to understand and predict the dynamics of antibiotic-sensitive mutants in a population of resistant bacteria during experimental evolution. Genetics 168 1131–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gelder, L., F. P. J. Vandecasteele, C. J. Brown, L. J. Forney and E. M. Top, 2005. Plasmid donor affects host range of the promiscuous IncP-1β plasmid pB10 in a sewage sludge microbial community. Appl. Environ. Microbiol. 71 5309–5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gelder, L., J. Ponciano, P. Joyce and E. M. Top, 2007. Stability of a promiscuous plasmid in different hosts: no guarantee for a long-term relationship. Microbiology 153 452–463. [DOI] [PubMed] [Google Scholar]
- de Visser, A. G. M., and D. E. Rozen, 2006. Clonal interference and the periodic selection of new beneficial mutations in Escherichia coli. Genetics 172 2093–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionisio, F., I. C. Conceicao, A. C. R. Marques, L. Fernandes and I. Gordo, 2005. The evolution of a conjugative plasmid and its ability to increase bacterial fitness. Biol. Lett. 1 250–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle, W. F., and E. Bapteste, 2007. Pattern pluralism and the Tree of Life hypothesis. Proc. Natl. Acad. Sci. USA 104 2043–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dröge, M., A. Pühler and W. Selbitschka, 1999. Horizontal gene transfer among bacteria in terrestrial and aquatic habitats as assessed by microcosm and field studies. Biol. Fertil. Soils 29 221–245. [Google Scholar]
- Dröge, M., A. Pühler and W. Selbitschka, 2000. Phenotypic and molecular characterization of conjugative antibiotic resistance plasmids isolated from bacterial communities of activated sludge. Mol. Gen. Genet. 263 471–482. [DOI] [PubMed] [Google Scholar]
- Duffy, S., C. L. Burch and P. E. Turner, 2007. Evolution of host specificity drives reproductive isolation among RNA viruses. Evol. Int. J. Org. Evol. 61 2614–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Tresguerres, M. E., M. Martin, D. Garcia de Viedma, R. Giraldo and R. Diaz-Orejas, 1995. Host growth temperature and a conservative amino acid substitution in the replication protein of pPS10 influence plasmid host range. J. Bacteriol. 177 4377–4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris, M. T., P. Joyce and C. L. Burch, 2007. High frequency of mutations that expand the host range of an RNA virus. Genetics 176 1013–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrish, P. J., and R. E. Lenski, 1998. The fate of competing beneficial mutations in an asexual population. Genetica 102/103 127–144. [PubMed] [Google Scholar]
- Gogarten, J. P., and J. P. Townsend, 2005. Horizontal gene transfer, genome innovation and evolution. Nat. Rev. 3 679–687. [DOI] [PubMed] [Google Scholar]
- Heuer, H., R. Fox and E. M. Top, 2007. Frequent conjugative transfer accelerates adaptation of an IncP-1 plasmid to an unfavourable Pseudomonas putida host. FEMS Microb. Ecol. 59 738–748. [DOI] [PubMed] [Google Scholar]
- Jain, R., M. C. Rivera, J. E. Moore and J. A. Lake, 2002. Horizontal gene transfer in microbial genome evolution. Theor. Pop. Biol. 61 489–495. [DOI] [PubMed] [Google Scholar]
- Koonin, E. V., 2003. Horizontal gene transfer: the path to maturity. Mol. Microb. 50 725–727. [DOI] [PubMed] [Google Scholar]
- Lawrence, J. R., and H. Hendrickson, 2003. Lateral gene transfer: When will adolescence end? Mol. Microb. 50 739–749. [DOI] [PubMed] [Google Scholar]
- Lenski, R. E., S. C. Simpson and T. T. Nguyen, 1994. Genetic analysis of a plasmid-encoded, host genotype-specific enhancement of bacterial fitness. J. Bacteriol. 176 3140–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévesque, C., S. Brassard, J. Lapointe and. P. H. Roy, 1994. Diversity and relative strength of tandem promoters for the antibiotic-resistance genes of several integrons. Gene 142 49–54. [DOI] [PubMed] [Google Scholar]
- Maestro, B., J. M. Sanz, M. Faelen, M. Couturier, R. Diaz-Orejas et al., 2002. Modulation of pPS10 host range by DnaA. Mol. Microbiol. 46 223–234. [DOI] [PubMed] [Google Scholar]
- Maestro, B., J. M. Sanz, R. Diaz-Orejas and E. Fernandez-Tresguerres, 2003. Modulation of pPS10 host range by plasmid-encoded RepA initiator protein. J. Bacteriol. 185 1367–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazodier, P., and J. Davies, 1991. Gene transfer between distantly related bacteria. Annu. Rev. Genet. 25 147–171. [DOI] [PubMed] [Google Scholar]
- McClure, N., A. Weightman and J. Fry, 1989. Survival of Pseudomonas putida UWC1 containing cloned catabolic genes in a model activated-sludge unit. Appl. Environ. Microbiol. 55 2627–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan, J. E., 2006. Resistance in nonfermenting gram-negative bacteria: multidrug resistance to the maximum. Am. J. Med. 119 S29–S36. [DOI] [PubMed] [Google Scholar]
- Modi, R. I., and J. Adams, 1991. Coevolution in bacterial-plasmid populations. Evolution 45 656–667. [DOI] [PubMed] [Google Scholar]
- Modi, R. I., C. M. Wilke, R. F. Rosenzweig and J. Adams, 1991. Plasmid macro-evolution: selection of deletions during adaptation in a nutrient-limited environment. Genetica 84 195–202. [DOI] [PubMed] [Google Scholar]
- Paterson, D. L., 2006. Resistance in gram-negative bacteria: Enterobacteriaceae. Am. J. Med. 119 S20–S28. [DOI] [PubMed] [Google Scholar]
- Ponciano, J. M., L. De Gelder, E. M. Top and P. Joyce, 2007. The population biology of bacterial plasmids: a hidden Markov model approach. Genetics 176 957–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlüter, A., H. Heuer, R. Szczepanowski, L. J. Forney, C. M. Thomas et al., 2003. The 64,508 bp IncP-1β antibiotic multiresistance plasmid pB10 isolated from a wastewater treatment plant provides evidence for recombination between members of different branches of the IncP-1B group. Microbiology 149 3139–3153. [DOI] [PubMed] [Google Scholar]
- Schlüter, A., R. Szczepanowski, A. Pühler and E. M. Top, 2007. Genomics of IncP-1 antibiotic resistance plasmids isolated from wastewater treatment plants provides evidence for a widely accessible drug resistance gene pool. FEMS Microbiol. Rev. 31 449–477. [DOI] [PubMed] [Google Scholar]
- Smets, B. F., and T. Barkay, 2005. Horizontal gene transfer: perspectives at a crossroads of scientific disciplines. Nat. Rev. Microbiol. 3 675–678. [DOI] [PubMed] [Google Scholar]
- Sota, M., and E. M. Top, 2008. Host-specific factors determine the persistence of IncP-1 plasmids. World J. Microbiol. Biotechnol. (in press).
- Sørensen, S. J., M. Bailey, L. H. Hansen, N. Kroer and S. Wuertz, 2005. Studying plasmid horizontal transfer in situ: a critical review. Nat. Rev. Microbiol. 3 700–710. [DOI] [PubMed] [Google Scholar]
- Stewart, F. M., and M. A. Levin, 1977. The population biology of bacterial plasmids: a priori conditions for the existence of conjugally transmitted factors. Genetics 87 209–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczepanowski, R., I. Krahn, A. Pühler and A. Schlüter, 2004. Different molecular rearrangements in the integron of the IncP-1 beta resistance plasmid pB10 isolated from a wastewater treatment plant result in elevated beta-lactam resistance levels. Arch. Microbiol. 182 429–435. [DOI] [PubMed] [Google Scholar]
- Thomas, C. M., 2000. The Horizontal Gene Pool. Bacterial Plasmids and Gene Spread. Harwood Academic Publishers, Amsterdam.
- Thomas, C. M., 2004. Evolution and population genetics of bacterial plasmids, pp. 509–528 in Plasmid Biology, edited by B. E. Funnell and G. J. Phillips. ASM Press, Washington, DC.
- Turner, P. E., V. S. Cooper and R. E. Lenski, 1998. Tradeoff between horizontal and vertical modes of transmission in bacterial plasmids. Evolution 52 315–329. [DOI] [PubMed] [Google Scholar]
- van Elsas, J. D., and M. J. Bailey, 2002. The ecology of transfer of mobile genetic elements. FEMS Microbiol. Ecol. 42 187–197. [DOI] [PubMed] [Google Scholar]
- Whitlock, M. C., 1996. The red queen beats the jack-of-all-trades: the limitations on the evolution of phenotypic plasticity and niche breadth. Am. Nat. 148 S65–S77. [Google Scholar]